Fkh1 and Fkh2 associate with Sir2 to control CLB2 transcription under normal and oxidative stress conditions
- PMID: 23874301
- PMCID: PMC3709100
- DOI: 10.3389/fphys.2013.00173
Fkh1 and Fkh2 associate with Sir2 to control CLB2 transcription under normal and oxidative stress conditions
Abstract
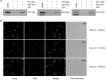
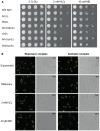
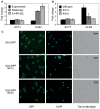
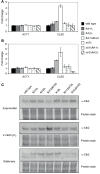
The Forkhead (Fkh) box family of transcription factors is evolutionary conserved from yeast to higher eukaryotes and its members are involved in many physiological processes including metabolism, DNA repair, cell cycle, stress resistance, apoptosis, and aging. In budding yeast, four Fkh transcription factors were identified, namely Fkh1, Fkh2, Fhl1, and Hcm1, which are implicated in chromatin silencing, cell cycle regulation, and stress response. These factors impinge transcriptional regulation during cell cycle progression, and histone deacetylases (HDACs) play an essential role in this process, e.g., the nuclear localization of Hcm1 depends on Sir2 activity, whereas Sin3/Rpd3 silence cell cycle specific gene transcription in G2/M phase. However, a direct involvement of Sir2 in Fkh1/Fkh2-dependent regulation of target genes is at present unknown. Here, we show that Fkh1 and Fkh2 associate with Sir2 in G1 and M phase, and that Fkh1/Fkh2-mediated activation of reporter genes is antagonized by Sir2. We further report that Sir2 overexpression strongly affects cell growth in an Fkh1/Fkh2-dependent manner. In addition, Sir2 regulates the expression of the mitotic cyclin Clb2 through Fkh1/Fkh2-mediated binding to the CLB2 promoter in G1 and M phase. We finally demonstrate that Sir2 is also enriched at the CLB2 promoter under stress conditions, and that the nuclear localization of Sir2 is dependent on Fkh1 and Fkh2. Taken together, our results show a functional interplay between Fkh1/Fkh2 and Sir2 suggesting a novel mechanism of cell cycle repression. Thus, in budding yeast, not only the regulation of G2/M gene expression but also the protective response against stress could be directly coordinated by Fkh1 and Fkh2.
Keywords: Fkh1; Fkh2; Sir2; budding yeast; cell cycle; silencing; stress.
Figures




Similar articles
-
New roles of DNA-binding and forkhead-associated domains of Fkh1 and Fkh2 in cellular functions.Cell Biochem Funct. 2022 Dec;40(8):888-902. doi: 10.1002/cbf.3750. Epub 2022 Sep 19. Cell Biochem Funct. 2022. PMID: 36121195
-
Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae.Genetics. 2000 Apr;154(4):1533-48. doi: 10.1093/genetics/154.4.1533. Genetics. 2000. PMID: 10747051 Free PMC article.
-
The Isw2 chromatin-remodeling ATPase cooperates with the Fkh2 transcription factor to repress transcription of the B-type cyclin gene CLB2.Mol Cell Biol. 2007 Apr;27(8):2848-60. doi: 10.1128/MCB.01798-06. Epub 2007 Feb 5. Mol Cell Biol. 2007. PMID: 17283050 Free PMC article.
-
Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes.Oncogene. 2005 Apr 18;24(17):2746-55. doi: 10.1038/sj.onc.1208606. Oncogene. 2005. PMID: 15838511 Review.
-
Histone acetylation and the cell-cycle in cancer.Front Biosci. 2001 Apr 1;6:D610-29. doi: 10.2741/1wang1. Front Biosci. 2001. PMID: 11282573 Review.
Cited by
-
Activating the Anaphase Promoting Complex to Enhance Genomic Stability and Prolong Lifespan.Int J Mol Sci. 2018 Jun 27;19(7):1888. doi: 10.3390/ijms19071888. Int J Mol Sci. 2018. PMID: 29954095 Free PMC article. Review.
-
A Clb/Cdk1-mediated regulation of Fkh2 synchronizes CLB expression in the budding yeast cell cycle.NPJ Syst Biol Appl. 2017 Mar 6;3:7. doi: 10.1038/s41540-017-0008-1. eCollection 2017. NPJ Syst Biol Appl. 2017. PMID: 28649434 Free PMC article.
-
Mitotic degradation of yeast Fkh1 by the Anaphase Promoting Complex is required for normal longevity, genomic stability and stress resistance.Aging (Albany NY). 2016 Apr;8(4):810-30. doi: 10.18632/aging.100949. Aging (Albany NY). 2016. PMID: 27099939 Free PMC article.
-
ChIP-exo analysis highlights Fkh1 and Fkh2 transcription factors as hubs that integrate multi-scale networks in budding yeast.Nucleic Acids Res. 2019 Sep 5;47(15):7825-7841. doi: 10.1093/nar/gkz603. Nucleic Acids Res. 2019. PMID: 31299083 Free PMC article.
-
Sirtuins-Mediated System-Level Regulation of Mammalian Tissues at the Interface between Metabolism and Cell Cycle: A Systematic Review.Biology (Basel). 2021 Mar 4;10(3):194. doi: 10.3390/biology10030194. Biology (Basel). 2021. PMID: 33806509 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

