The PyRosetta Toolkit: a graphical user interface for the Rosetta software suite
- PMID: 23874400
- PMCID: PMC3706480
- DOI: 10.1371/journal.pone.0066856
The PyRosetta Toolkit: a graphical user interface for the Rosetta software suite
Abstract
The Rosetta Molecular Modeling suite is a command-line-only collection of applications that enable high-resolution modeling and design of proteins and other molecules. Although extremely useful, Rosetta can be difficult to learn for scientists with little computational or programming experience. To that end, we have created a Graphical User Interface (GUI) for Rosetta, called the PyRosetta Toolkit, for creating and running protocols in Rosetta for common molecular modeling and protein design tasks and for analyzing the results of Rosetta calculations. The program is highly extensible so that developers can add new protocols and analysis tools to the PyRosetta Toolkit GUI.
Conflict of interest statement
Figures
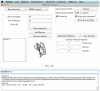
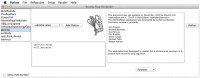
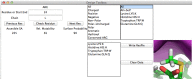
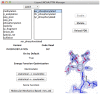
References
-
- Das R, Baker D (2008) Macromolecular modeling with rosetta. Ann. Rev. Biochem. 77: 363–382. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

