Still acting green: continued expression of photosynthetic genes in the heterotrophic Dinoflagellate Pfiesteria piscicida (Peridiniales, Alveolata)
- PMID: 23874554
- PMCID: PMC3712967
- DOI: 10.1371/journal.pone.0068232
Still acting green: continued expression of photosynthetic genes in the heterotrophic Dinoflagellate Pfiesteria piscicida (Peridiniales, Alveolata)
Abstract
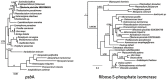
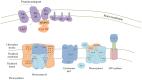
The loss of photosynthetic function should lead to the cessation of expression and finally loss of photosynthetic genes in the new heterotroph. Dinoflagellates are known to have lost their photosynthetic ability several times. Dinoflagellates have also acquired photosynthesis from other organisms, either on a long-term basis or as "kleptoplastids" multiple times. The fate of photosynthetic gene expression in heterotrophs can be informative into evolution of gene expression patterns after functional loss, and the dinoflagellates ability to acquire new photosynthetic function through additional endosymbiosis. To explore this we analyzed a large-scale EST database consisting of 151,091 unique sequences (29,170 contigs, 120,921 singletons) obtained from 454 pyrosequencing of the heterotrophic dinoflagellate Pfiesteria piscicida. About 597 contigs from P. piscicida showed significant homology (E-value <e(-30)) with proteins associated with plastid and photosynthetic function. Most of the genes involved in the Calvin-Benson cycle were found, genes of the light-dependent reaction were also identified. Also genes of associated pathways including the chorismate pathway and genes involved in starch metabolism were discovered. BLAST searches and phylogenetic analysis suggest that these plastid-associated genes originated from several different photosynthetic ancestors. The Calvin-Benson cycle genes are mostly associated with genes derived from the secondary plastids of peridinin-containing dinoflagellates, while the light-harvesting genes are derived from diatoms, or diatoms that are tertiary plastids in other dinoflagellates. The continued expression of many genes involved in photosynthetic pathways indicates that the loss of transcriptional regulation may occur well after plastid loss and could explain the organism's ability to "capture" new plastids (i.e. different secondary endosymbiosis or tertiary symbioses) to renew photosynthetic function.
Conflict of interest statement
Figures



Similar articles
-
Plastid genes in a non-photosynthetic dinoflagellate.Protist. 2007 Jan;158(1):105-17. doi: 10.1016/j.protis.2006.09.004. Epub 2006 Dec 5. Protist. 2007. PMID: 17150410
-
Investigation of heterotrophs reveals new insights in dinoflagellate evolution.Mol Phylogenet Evol. 2024 Jul;196:108086. doi: 10.1016/j.ympev.2024.108086. Epub 2024 Apr 26. Mol Phylogenet Evol. 2024. PMID: 38677354
-
Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements.J Mol Evol. 2001 Sep;53(3):204-13. doi: 10.1007/s002390010210. J Mol Evol. 2001. PMID: 11523007
-
Integration of plastids with their hosts: Lessons learned from dinoflagellates.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10247-54. doi: 10.1073/pnas.1421380112. Epub 2015 May 20. Proc Natl Acad Sci U S A. 2015. PMID: 25995366 Free PMC article. Review.
-
The endosymbiotic origin, diversification and fate of plastids.Philos Trans R Soc Lond B Biol Sci. 2010 Mar 12;365(1541):729-48. doi: 10.1098/rstb.2009.0103. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20124341 Free PMC article. Review.
Cited by
-
Identification of three proteins involved in fertilization and parthenogenetic development of a brown alga, Scytosiphon lomentaria.Planta. 2014 Dec;240(6):1253-67. doi: 10.1007/s00425-014-2148-5. Epub 2014 Aug 21. Planta. 2014. PMID: 25143248
-
The Cryptic Plastid of Euglena longa Defines a New Type of Nonphotosynthetic Plastid Organelle.mSphere. 2020 Oct 21;5(5):e00675-20. doi: 10.1128/mSphere.00675-20. mSphere. 2020. PMID: 33087518 Free PMC article.
-
Genes functioned in kleptoplastids of Dinophysis are derived from haptophytes rather than from cryptophytes.Sci Rep. 2019 Jun 21;9(1):9009. doi: 10.1038/s41598-019-45326-5. Sci Rep. 2019. PMID: 31227737 Free PMC article.
-
Reduced plastid genomes of colorless facultative pathogens Prototheca (Chlorophyta) are retained for membrane transport genes.BMC Biol. 2024 Dec 18;22(1):294. doi: 10.1186/s12915-024-02089-4. BMC Biol. 2024. PMID: 39696433 Free PMC article.
-
Feeding diverse prey as an excellent strategy of mixotrophic dinoflagellates for global dominance.Sci Adv. 2021 Jan 8;7(2):eabe4214. doi: 10.1126/sciadv.abe4214. Print 2021 Jan. Sci Adv. 2021. PMID: 33523999 Free PMC article.
References
-
- Fast NM, Kissinger JC, Roos DS, Keeling PJ (2001) Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol Biol Evol 18: 418–426. - PubMed
-
- Archibald JM (2009) The puzzle of plastid evolution. Curr Biol 19: R81–R88. - PubMed
-
- Johnson MD (2011) The acquisition of phototrophy: Adaptive strategies of hosting endosymbionts and organelles. Photosynth Res 107: 117–132. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

