Super resolution microscopy reveals that caveolin-1 is required for spatial organization of CRFB1 and subsequent antiviral signaling in zebrafish
- PMID: 23874753
- PMCID: PMC3706321
- DOI: 10.1371/journal.pone.0068759
Super resolution microscopy reveals that caveolin-1 is required for spatial organization of CRFB1 and subsequent antiviral signaling in zebrafish
Abstract
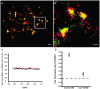
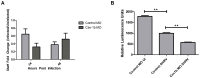
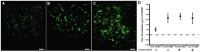
Understanding spatial distribution and dynamics of receptors within unperturbed membranes is essential for elucidating their role in antiviral signaling, but conventional studies of detergent-resistant membrane fractions cannot provide this information. Caveolae are integral to numerous signaling pathways and these membrane domains have been previously implicated in viral entry but not antiviral defense. This study shows, for the first time, the importance of spatio-temporal regulation of signaling receptors and the importance of the regulation of clustering for downstream signaling. A novel mechanism for virus evasion of host cell defenses is demonstrated through disruption of clusters of signaling molecules organized within caveolin-rich domains. Viral infection leads to a downregulation in Caveolin-1b (Cav-1b), disrupting clusters of CRFB1, a zebrafish type I interferon receptor (-R) subunit. Super-resolution microscopy has enabled the first single-molecule imaging of CRFB1 association with cav-1b-containing membrane domains. Strikingly, downregulation of Cav-1b, the major protein component of caveolae, caused CRFB1 clusters to disperse. Dispersal of CRFB1 clusters led to a suppressed antiviral immune response both in vitro and in vivo, through abrogation of downstream signaling. This response strongly suggests that CRFB1 organization within cav-1b-containing membrane domains is critical for IFN-mediated antiviral defense and presents a previously undescribed antiviral evasion strategy to alter IFN signaling and the antiviral immune response.
Conflict of interest statement
Figures


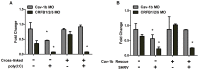
References
-
- Okamoto T, Schlegel A, Scherer PE, Lisanti MP (1998) Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem 273: 5419–5422. - PubMed
-
- Lisanti MP, Scherer PE, Tang Z, Sargiacomo M (1994) Caveolae, caveolin and caveolin-rich membrane domains: a signalling hypothesis. Trends Cell Biol 4: 231–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

