Oral neutrophil transcriptome changes result in a pro-survival phenotype in periodontal diseases
- PMID: 23874838
- PMCID: PMC3708893
- DOI: 10.1371/journal.pone.0068983
Oral neutrophil transcriptome changes result in a pro-survival phenotype in periodontal diseases
Abstract
Background: Periodontal diseases are inflammatory processes that occur following the influx of neutrophils into the periodontal tissues in response to the subgingival bacterial biofilm. Current literature suggests that while neutrophils are protective and prevent bacterial infections, they also appear to contribute to damage of the periodontal tissues. In the present study we compare the gene expression profile changes in neutrophils as they migrate from the circulation into the oral tissues in patients with chronic periodontits and matched healthy subjects. We hypothesized that oral neutrophils in periodontal disease patients will display a disease specific transcriptome that differs from the oral neutrophil of healthy subjects.
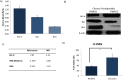
Methods: Venous blood and oral rinse samples were obtained from healthy subjects and chronic periodontitis patients for neutrophil isolation. mRNA was isolated from the neutrophils, and gene expression microarray analysis was completed. Results were confirmed for specific genes of interest by qRT-PCR and Western Blot analysis.
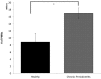
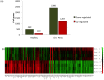
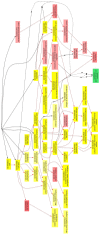
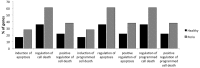
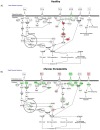
Results and discussion: Chronic periodontitis patients presented with increased recruitment of neutrophils to the oral cavity. Gene expression analysis revealed differences in the expression levels of genes from several biological pathways. Using hierarchical clustering analysis, we found that the apoptosis network was significantly altered in patients with chronic inflammation in the oral cavity, with up-regulation of pro-survival members of the Bcl-2 family and down-regulation of pro-apoptosis members in the same compartment. Additional functional analysis confirmed that the percentages of viable neutrophils are significantly increased in the oral cavity of chronic periodontitis patients.
Conclusions: Oral neutrophils from patients with periodontal disease displayed an altered transcriptome following migration into the oral tissues. This resulted in a pro-survival neutrophil phenotype in chronic periodontitis patients when compared with healthy subjects, resulting in a longer-lived neutrophil. This is likely to impact the severity and length of the inflammatory response in this oral disease.
Conflict of interest statement
Figures

References
-
- Graves D (2008) Cytokines that promote periodontal tissue destruction. Journal of Periodontology 79: 1585–1591 doi:10.1902/jop.2008.080183 - DOI - PubMed
-
- Guentsch A, Puklo M, Preshaw PM, Glockmann E, Pfister W, et al. (2009) Neutrophils in chronic and aggressive periodontitis in interaction with Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans. J Periodontal Res 44: 368–377 doi:10.1111/j.1600-0765.2008.01113.x - DOI - PMC - PubMed
-
- Taba M Jr, Kinney J, Kim AS, Giannobile WV (2005) Diagnostic Biomarkers for Oral and Periodontal Diseases. Dental Clinics of North America 49: 551–571 doi:10.1016/j.cden.2005.03.009 - DOI - PMC - PubMed
-
- Offenbacher S, Barros SP, Beck JD (2008) Rethinking Periodontal Inflammation. Journal of Periodontology 79: 1577–1584 doi:10.1902/jop.2008.080220 - DOI - PubMed
-
- Hernández M, Gamonal J, Tervahartiala T, Mäntylä P, Rivera O, et al. (2010) Associations Between Matrix Metalloproteinase-8 and -14 and Myeloperoxidase in Gingival Crevicular Fluid From Subjects With Progressive Chronic Periodontitis: A Longitudinal Study. Journal of Periodontology 81: 1644–1652 doi:10.1902/jop.2010.100196 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

