Deletion of Snai2 and Snai3 results in impaired physical development compounded by lymphocyte deficiency
- PMID: 23874916
- PMCID: PMC3713067
- DOI: 10.1371/journal.pone.0069216
Deletion of Snai2 and Snai3 results in impaired physical development compounded by lymphocyte deficiency
Abstract
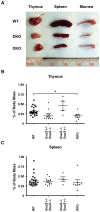
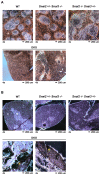
The Snail family of transcriptional regulators consists of three highly conserved members. These proteins regulate (repress) transcription via the recruitment of histone deacetylases to target gene promoters that possess the appropriate E-box binding sequences. Murine Snai1 is required for mouse development while Snai2 deficient animals survive with some anomalies. Less is known about the third member of the family, Snai3. To investigate the function of Snai3, we generated a conditional knockin mouse. Utilizing Cre-mediated deletion to facilitate the ablation of Snai3 in T cells or the entire animal, we found little to no effect of the loss of Snai3 in the entire animal or in T cell lineages. This finding provided the hypothesis that absence of Snai3 was mitigated, in part, by the presence of Snai2. To test this hypothesis we created Snai2/Snai3 double deficient mice. The developmental consequences of lacking both of these proteins was manifested in stunted growth, a paucity of offspring including a dramatic deficiency of female mice, and impaired immune cell development within the lymphoid lineages.
Conflict of interest statement
Figures









Similar articles
-
Fatal autoimmunity results from the conditional deletion of Snai2 and Snai3.Cell Immunol. 2015 May;295(1):1-18. doi: 10.1016/j.cellimm.2015.02.009. Epub 2015 Feb 24. Cell Immunol. 2015. PMID: 25732600 Free PMC article.
-
Snai2 and Snai3 transcriptionally regulate cellular fitness and functionality of T cell lineages through distinct gene programs.Immunobiology. 2016 May;221(5):618-33. doi: 10.1016/j.imbio.2016.01.007. Epub 2016 Jan 22. Immunobiology. 2016. PMID: 26831822 Free PMC article.
-
The snail family gene snai3 is not essential for embryogenesis in mice.PLoS One. 2013 Jun 6;8(6):e65344. doi: 10.1371/journal.pone.0065344. Print 2013. PLoS One. 2013. PMID: 23762348 Free PMC article.
-
Snail transcription factors in hematopoietic cell development: a model of functional redundancy.Exp Hematol. 2014 Jun;42(6):425-30. doi: 10.1016/j.exphem.2014.03.002. Epub 2014 Mar 24. Exp Hematol. 2014. PMID: 24674754 Free PMC article. Review.
-
WNT signaling and lymphocyte development.Cell. 2002 Apr;109 Suppl:S13-9. doi: 10.1016/s0092-8674(02)00709-2. Cell. 2002. PMID: 11983149 Review.
Cited by
-
Fatal autoimmunity results from the conditional deletion of Snai2 and Snai3.Cell Immunol. 2015 May;295(1):1-18. doi: 10.1016/j.cellimm.2015.02.009. Epub 2015 Feb 24. Cell Immunol. 2015. PMID: 25732600 Free PMC article.
-
Prognostic values of SNAI family members in breast cancer patients.Ann Transl Med. 2020 Aug;8(15):922. doi: 10.21037/atm-20-681. Ann Transl Med. 2020. PMID: 32953722 Free PMC article.
-
Snai2 and Snai3 transcriptionally regulate cellular fitness and functionality of T cell lineages through distinct gene programs.Immunobiology. 2016 May;221(5):618-33. doi: 10.1016/j.imbio.2016.01.007. Epub 2016 Jan 22. Immunobiology. 2016. PMID: 26831822 Free PMC article.
-
An "unexpected" role for EMT transcription factors in hematological development and malignancy.Front Immunol. 2023 Aug 3;14:1207360. doi: 10.3389/fimmu.2023.1207360. eCollection 2023. Front Immunol. 2023. PMID: 37600794 Free PMC article. Review.
-
Evidence from a mouse model that epithelial cell migration and mesenchymal-epithelial transition contribute to rapid restoration of uterine tissue integrity during menstruation.PLoS One. 2014 Jan 22;9(1):e86378. doi: 10.1371/journal.pone.0086378. eCollection 2014. PLoS One. 2014. PMID: 24466063 Free PMC article.
References
-
- Barrallo-Gimeno A, Nieto MA (2009) Evolutionary history of the Snail/Scratch superfamily. Trends Genet 25: 248-252. doi:10.1016/j.tig.2009.04.001. PubMed: 19427053. - DOI - PubMed
-
- Manzanares M, Blanco MJ, Nieto MA (2004) Snail3 orthologues in vertebrates: divergent members of the Snail zinc-finger gene family. Dev Genes Evol 214: 47-53. doi:10.1007/s00427-003-0373-1. PubMed: 14655014. - DOI - PubMed
-
- Boulay JL, Dennefeld C, Alberga A (1987) The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature 330: 395-398. doi:10.1038/330395a0. PubMed: 3683556. - DOI - PubMed
-
- Nieto MA (2002) The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol 3: 155-166. doi:10.1038/nrm757. PubMed: 11994736. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

