Turnover of amyloid precursor protein family members determines their nuclear signaling capability
- PMID: 23874953
- PMCID: PMC3715505
- DOI: 10.1371/journal.pone.0069363
Turnover of amyloid precursor protein family members determines their nuclear signaling capability
Abstract
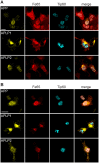
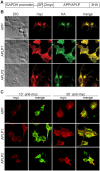
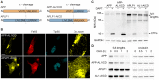
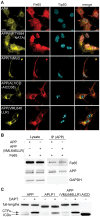
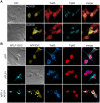
The amyloid precursor protein (APP) as well as its homologues, APP-like protein 1 and 2 (APLP1 and APLP2), are cleaved by α-, β-, and γ-secretases, resulting in the release of their intracellular domains (ICDs). We have shown that the APP intracellular domain (AICD) is transported to the nucleus by Fe65 where they jointly bind the histone acetyltransferase Tip60 and localize to spherical nuclear complexes (AFT complexes), which are thought to be sites of transcription. We have now analyzed the subcellular localization and turnover of the APP family members. Similarly to AICD, the ICD of APLP2 localizes to spherical nuclear complexes together with Fe65 and Tip60. In contrast, the ICD of APLP1, despite binding to Fe65, does not translocate to the nucleus. In addition, APLP1 predominantly localizes to the plasma membrane, whereas APP and APLP2 are detected in vesicular structures. APLP1 also demonstrates a much slower turnover of the full-length protein compared to APP and APLP2. We further show that the ICDs of all APP family members are degraded by the proteasome and that the N-terminal amino acids of ICDs determine ICD degradation rate. Together, our results suggest that different nuclear signaling capabilities of APP family members are due to different rates of full-length protein processing and ICD proteasomal degradation. Our results provide evidence in support of a common nuclear signaling function for APP and APLP2 that is absent in APLP1, but suggest that APLP1 has a regulatory role in the nuclear translocation of APP family ICDs due to the sequestration of Fe65.
Conflict of interest statement
Figures



References
-
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, et al. (1987) The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature 325: 733–736. - PubMed
-
- De Strooper B, Annaert W (2000) Proteolytic processing and cell biological functions of the amyloid precursor protein. J Cell Sci 113 (Pt 11): 1857–1870. - PubMed
-
- Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297: 353–356. - PubMed
-
- von Rotz RC, Kohli BM, Bosset J, Meier M, Suzuki T, et al. (2004) The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J Cell Sci 117: 4435–4448. - PubMed
-
- Weidemann A, Eggert S, Reinhard FB, Vogel M, Paliga K, et al. (2002) A novel epsilon-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with Notch processing. Biochemistry 41: 2825–2835. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

