A lover and a fighter: the genome sequence of an entomopathogenic nematode Heterorhabditis bacteriophora
- PMID: 23874975
- PMCID: PMC3715494
- DOI: 10.1371/journal.pone.0069618
A lover and a fighter: the genome sequence of an entomopathogenic nematode Heterorhabditis bacteriophora
Abstract
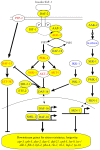
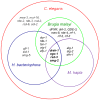
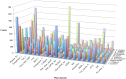
Heterorhabditis bacteriophora are entomopathogenic nematodes that have evolved a mutualism with Photorhabdus luminescens bacteria to function as highly virulent insect pathogens. The nematode provides a safe harbor for intestinal symbionts in soil and delivers the symbiotic bacteria into the insect blood. The symbiont provides virulence and toxins, metabolites essential for nematode reproduction, and antibiotic preservation of the insect cadaver. Approximately half of the 21,250 putative protein coding genes identified in the 77 Mbp high quality draft H. bacteriophora genome sequence were novel proteins of unknown function lacking homologs in Caenorhabditis elegans or any other sequenced organisms. Similarly, 317 of the 603 predicted secreted proteins are novel with unknown function in addition to 19 putative peptidases, 9 peptidase inhibitors and 7 C-type lectins that may function in interactions with insect hosts or bacterial symbionts. The 134 proteins contained mariner transposase domains, of which there are none in C. elegans, suggesting an invasion and expansion of mariner transposons in H. bacteriophora. Fewer Kyoto Encyclopedia of Genes and Genomes Orthologies in almost all metabolic categories were detected in the genome compared with 9 other sequenced nematode genomes, which may reflect dependence on the symbiont or insect host for these functions. The H. bacteriophora genome sequence will greatly facilitate genetics, genomics and evolutionary studies to gain fundamental knowledge of nematode parasitism and mutualism. It also elevates the utility of H. bacteriophora as a bridge species between vertebrate parasitic nematodes and the C. elegans model.
Conflict of interest statement
Figures


References
-
- Bongers T, Ferris H (1999) Nematode community structure as a bioindicator in environmental monitoring. Trends in Ecology and Evolution 14: 224–228. - PubMed
-
- Baldwin JG, Nadler SA, Adams BJ (2004) Evolution of plant parasitism among nematodes. Annu Rev Phytopathol 42: 83–105. - PubMed
-
- Gaugler R (2002) Entomopathogenic nematology. Wallingford, UK: CABI Publishing.
-
- Grewal PS, Ehlers RU, Shapiro-Ilan DI (2005) Nematodes as biocontrol agents. Wallingford, UK: CABI Publishing. 480 p.
-
- De Ley P, Blaxter M (2002) Systematic position and phylogeny. In: Lee DL, editor. The Biology of Nematodes. London, UK: Taylor and Francis. 1–30.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

