The advantages and limitations of trait analysis with GWAS: a review
- PMID: 23876160
- PMCID: PMC3750305
- DOI: 10.1186/1746-4811-9-29
The advantages and limitations of trait analysis with GWAS: a review
Abstract
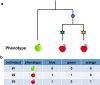
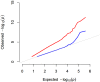
Over the last 10 years, high-density SNP arrays and DNA re-sequencing have illuminated the majority of the genotypic space for a number of organisms, including humans, maize, rice and Arabidopsis. For any researcher willing to define and score a phenotype across many individuals, Genome Wide Association Studies (GWAS) present a powerful tool to reconnect this trait back to its underlying genetics. In this review we discuss the biological and statistical considerations that underpin a successful analysis or otherwise. The relevance of biological factors including effect size, sample size, genetic heterogeneity, genomic confounding, linkage disequilibrium and spurious association, and statistical tools to account for these are presented. GWAS can offer a valuable first insight into trait architecture or candidate loci for subsequent validation.
Keywords: Arabidopsis; Effect size; GWAS; Genetic heterogeneity; Mixed model.
Figures


References
-
- Kowalski SP, Lan TH, Feldmann KA, Paterson AH. QTL mapping of naturally-occurring variation in flowering time of Arabidopsis thaliana. Mol Gen Genet. 1994;245(5):548–555. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

