VlincRNAs controlled by retroviral elements are a hallmark of pluripotency and cancer
- PMID: 23876380
- PMCID: PMC4053963
- DOI: 10.1186/gb-2013-14-7-r73
VlincRNAs controlled by retroviral elements are a hallmark of pluripotency and cancer
Abstract
Background: The function of the non-coding portion of the human genome remains one of the most important questions of our time. Its vast complexity is exemplified by the recent identification of an unusual and notable component of the transcriptome - very long intergenic non-coding RNAs, termed vlincRNAs.
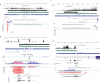
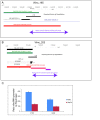
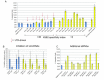
Results: Here we identify 2,147 vlincRNAs covering 10 percent of our genome. We show they are present not only in cancerous cells, but also in primary cells and normal human tissues, and are controlled by canonical promoters. Furthermore, vlincRNA promoters frequently originate from within endogenous retroviral sequences. Strikingly, the number of vlincRNAs expressed from endogenous retroviral promoters strongly correlates with pluripotency or the degree of malignant transformation. These results suggest a previously unknown connection between the pluripotent state and cancer via retroviral repeat-driven expression of vlincRNAs. Finally, we show that vlincRNAs can be syntenically conserved in humans and mouse and their depletion using RNAi can cause apoptosis in cancerous cells.
Conclusions: These intriguing observations suggest that vlincRNAs could create a framework that combines many existing short ESTs and lincRNAs into a landscape of very long transcripts functioning in the regulation of gene expression in the nucleus. Certain types of vlincRNAs participate at specific stages of normal development and, based on analysis of a limited set of cancerous and primary cell lines, they appear to be co-opted by cancer-associated transcriptional programs. This provides additional understanding of transcriptome regulation during the malignant state, and could lead to additional targets and options for its reversal.
Figures



References
-
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, Rozowsky JS, Gerstein MB, Wahlestedt C, Hayashizaki Y, Carninci P, Gingeras TR, Mattick JS. The reality of pervasive transcription. PLoS Biol. 2011;14:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

