The ribosome triggers the stringent response by RelA via a highly distorted tRNA
- PMID: 23877429
- PMCID: PMC3790049
- DOI: 10.1038/embor.2013.106
The ribosome triggers the stringent response by RelA via a highly distorted tRNA
Abstract
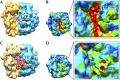
The bacterial stringent response links nutrient starvation with the transcriptional control of genes. This process is initiated by the stringent factor RelA, which senses the presence of deacylated tRNA in the ribosome as a symptom of amino-acid starvation to synthesize the alarmone (p)ppGpp. Here we report a cryo-EM study of RelA bound to ribosomes bearing cognate, deacylated tRNA in the A-site. The data show that RelA on the ribosome stabilizes an unusual distorted form of the tRNA, with the acceptor arm making contact with RelA and far from its normal location in the peptidyl transferase centre.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

