Structures of the class D Carbapenemases OXA-23 and OXA-146: mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins, and aztreonam
- PMID: 23877677
- PMCID: PMC3811470
- DOI: 10.1128/AAC.00762-13
Structures of the class D Carbapenemases OXA-23 and OXA-146: mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins, and aztreonam
Abstract
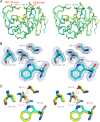
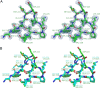
Class D β-lactamases that hydrolyze carbapenems such as imipenem and doripenem are a recognized danger to the efficacy of these "last-resort" β-lactam antibiotics. Like all known class D carbapenemases, OXA-23 cannot hydrolyze the expanded-spectrum cephalosporin ceftazidime. OXA-146 is an OXA-23 subfamily clinical variant that differs from the parent enzyme by a single alanine (A220) inserted in the loop connecting β-strands β5 and β6. We discovered that this insertion enables OXA-146 to bind and hydrolyze ceftazidime with an efficiency comparable to those of other extended-spectrum class D β-lactamases. OXA-146 also binds and hydrolyzes aztreonam, cefotaxime, ceftriaxone, and ampicillin with higher efficiency than OXA-23 and preserves activity against doripenem. In this study, we report the X-ray crystal structures of both the OXA-23 and OXA-146 enzymes at 1.6-Å and 1.2-Å resolution. A comparison of the two structures shows that the extra alanine moves a methionine (M221) out of its normal position, where it forms a bridge over the top of the active site. This single amino acid insertion also lengthens the β5-β6 loop, moving the entire backbone of this region further away from the active site. A model of ceftazidime bound in the active site reveals that these two structural alterations are both likely to relieve steric clashes between the bulky R1 side chain of ceftazidime and OXA-23. With activity against all four classes of β-lactam antibiotics, OXA-146 represents an alarming new threat to the treatment of infections caused by Acinetobacter spp.
Figures



References
-
- Naas T, Poirel L, Nordmann P. 2008. Minor extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14(Suppl 1):42–52 - PubMed
-
- Walther-Rasmussen J, Hoiby N. 2006. OXA-type carbapenemases. J. Antimicrob. Chemother. 57:373–383 - PubMed
-
- Scaife W, Young HK, Paton RH, Amyes SG. 1995. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J. Antimicrob. Chemother. 36:585–586 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

