Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease
- PMID: 23878218
- PMCID: PMC3740842
- DOI: 10.1073/pnas.1311861110
Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease
Abstract
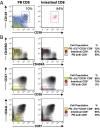
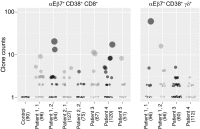
Celiac disease is an intestinal autoimmune disease driven by dietary gluten and gluten-specific CD4(+) T-cell responses. In celiac patients on a gluten-free diet, exposure to gluten induces the appearance of gluten-specific CD4(+) T cells with gut-homing potential in the peripheral blood. Here we show that gluten exposure also induces the appearance of activated, gut-homing CD8(+) αβ and γδ T cells in the peripheral blood. Single-cell T-cell receptor sequence analysis indicates that both of these cell populations have highly focused T-cell receptor repertoires, indicating that their induction is antigen-driven. These results reveal a previously unappreciated role of antigen in the induction of CD8(+) αβ and γδ T cells in celiac disease and demonstrate a coordinated response by all three of the major types of T cells. More broadly, these responses may parallel adaptive immune responses to viral pathogens and other systemic autoimmune diseases.
Keywords: autoimmunity; mucosal immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Fallang LE, et al. Differences in the risk of celiac disease associated with HLA-DQ2.5 or HLA-DQ2.2 are related to sustained gluten antigen presentation. Nat Immunol. 2009;10(10):1096–1101. - PubMed
-
- Brottveit M, et al. Assessing possible celiac disease by an HLA-DQ2-gliadin tetramer test. Am J Gastroenterol. 2011;106(7):1318–1324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

