Capturing the mutational landscape of the beta-lactamase TEM-1
- PMID: 23878237
- PMCID: PMC3740883
- DOI: 10.1073/pnas.1215206110
Capturing the mutational landscape of the beta-lactamase TEM-1
Abstract
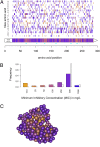
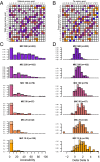
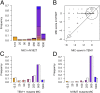
Adaptation proceeds through the selection of mutations. The distribution of mutant fitness effect and the forces shaping this distribution are therefore keys to predict the evolutionary fate of organisms and their constituents such as enzymes. Here, by producing and sequencing a comprehensive collection of 10,000 mutants, we explore the mutational landscape of one enzyme involved in the spread of antibiotic resistance, the beta-lactamase TEM-1. We measured mutation impact on the enzyme activity through the estimation of amoxicillin minimum inhibitory concentration on a subset of 990 mutants carrying a unique missense mutation, representing 64% of possible amino acid changes in that protein reachable by point mutation. We established that mutation type, solvent accessibility of residues, and the predicted effect of mutations on protein stability primarily determined alone or in combination changes in minimum inhibitory concentration of mutants. Moreover, we were able to capture the drastic modification of the mutational landscape induced by a single stabilizing point mutation (M182T) by a simple model of protein stability. This work thereby provides an integrated framework to study mutation effects and a tool to understand/define better the epistatic interactions.
Keywords: adaptive landscape; distribution of fitness effects; epistasis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

