Rapid and robust signaling in the CsrA cascade via RNA-protein interactions and feedback regulation
- PMID: 23878244
- PMCID: PMC3740896
- DOI: 10.1073/pnas.1308476110
Rapid and robust signaling in the CsrA cascade via RNA-protein interactions and feedback regulation
Abstract
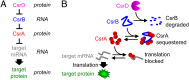
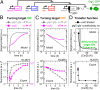
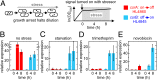
Bacterial survival requires the rapid propagation of signals through gene networks during stress, but how this is achieved is not well understood. This study systematically characterizes the signaling dynamics of a cascade of RNA-protein interactions in the CsrA system, which regulates stress responses and biofilm formation in Escherichia coli. Noncoding RNAs are at the center of the CsrA system; target mRNAs are bound by CsrA proteins that inhibit their translation, CsrA proteins are sequestered by CsrB noncoding RNAs, and the degradation of CsrB RNAs is increased by CsrD proteins. Here, we show using in vivo experiments and quantitative modeling that the CsrA system integrates three strategies to achieve rapid and robust signaling. These strategies include: (i) the sequestration of stable proteins by noncoding RNAs, which rapidly inactivates protein activity; (ii) the degradation of stable noncoding RNAs, which enables their rapid removal; and (iii) a negative-feedback loop created by CsrA repression of CsrD production, which reduces the time for the system to achieve steady state. We also demonstrate that sequestration in the CsrA system results in signaling that is robust to growth rates because it does not rely on the slow dilution of molecules via cell division; therefore, signaling can occur even during growth arrest induced by starvation or antibiotic treatment.
Keywords: gene circuit; reverse engineering; synthetic biology; systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J Mol Biol. 2002;323(5):785–793. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

