Inhibition of Cdc42 during mitotic exit is required for cytokinesis
- PMID: 23878274
- PMCID: PMC3718968
- DOI: 10.1083/jcb.201301090
Inhibition of Cdc42 during mitotic exit is required for cytokinesis
Abstract
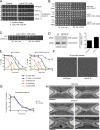
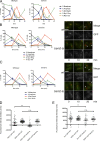
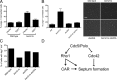
The role of Cdc42 and its regulation during cytokinesis is not well understood. Using biochemical and imaging approaches in budding yeast, we demonstrate that Cdc42 activation peaks during the G1/S transition and during anaphase but drops during mitotic exit and cytokinesis. Cdc5/Polo kinase is an important upstream cell cycle regulator that suppresses Cdc42 activity. Failure to down-regulate Cdc42 during mitotic exit impairs the normal localization of key cytokinesis regulators-Iqg1 and Inn1-at the division site, and results in an abnormal septum. The effects of Cdc42 hyperactivation are largely mediated by the Cdc42 effector p21-activated kinase Ste20. Inhibition of Cdc42 and related Rho guanosine triphosphatases may be a general feature of cytokinesis in eukaryotes.
Figures


Comment in
-
Timing it right: precise ON/OFF switches for Rho1 and Cdc42 GTPases in cytokinesis.J Cell Biol. 2013 Jul 22;202(2):187-9. doi: 10.1083/jcb.201306152. J Cell Biol. 2013. PMID: 23878271 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

