Integrative genomic analysis of the human immune response to influenza vaccination
- PMID: 23878721
- PMCID: PMC3713456
- DOI: 10.7554/eLife.00299
Integrative genomic analysis of the human immune response to influenza vaccination
Erratum in
-
Correction: Integrative genomic analysis of the human immune response to influenza vaccination.Elife. 2016 Aug 30;5:e18898. doi: 10.7554/eLife.18898. Elife. 2016. PMID: 27576045 Free PMC article. No abstract available.
Abstract
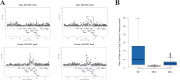
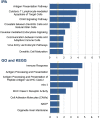
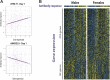
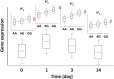
Identification of the host genetic factors that contribute to variation in vaccine responsiveness may uncover important mechanisms affecting vaccine efficacy. We carried out an integrative, longitudinal study combining genetic, transcriptional, and immunologic data in humans given seasonal influenza vaccine. We identified 20 genes exhibiting a transcriptional response to vaccination, significant genotype effects on gene expression, and correlation between the transcriptional and antibody responses. The results show that variation at the level of genes involved in membrane trafficking and antigen processing significantly influences the human response to influenza vaccination. More broadly, we demonstrate that an integrative study design is an efficient alternative to existing methods for the identification of genes involved in complex traits. DOI:http://dx.doi.org/10.7554/eLife.00299.001.
Keywords: Complex-trait genetics; Human; Human genetics; Integrative biology; Systems biology; Vaccines; eQTL.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




Comment in
-
Immune response is a personal matter.Elife. 2013 Jul 16;2:e00899. doi: 10.7554/eLife.00899. Elife. 2013. PMID: 23878727 Free PMC article.
References
-
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, Schubert LA, Birditt B, Shay T, Goren A, Zhang X, Smith Z, Deering R, McDonald RC, Cabili M, Bernstein BE, Rinn JL, Meissner A, Root DE, Hacohen N, Regev A. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. - DOI - PMC - PubMed
-
- Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. - DOI - PMC - PubMed
-
- Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Niño D, Arden N, Quarles JM, Couch RB, Belmont JW. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. Journal of Infectious Diseases. 2011;203:921–929. doi: 10.1093/infdis/jiq156. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

