Emerging roles of the nucleolus in regulating the DNA damage response: the noncanonical DNA repair enzyme APE1/Ref-1 as a paradigmatical example
- PMID: 23879289
- PMCID: PMC3901381
- DOI: 10.1089/ars.2013.5491
Emerging roles of the nucleolus in regulating the DNA damage response: the noncanonical DNA repair enzyme APE1/Ref-1 as a paradigmatical example
Abstract
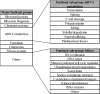
Significance: An emerging concept in DNA repair mechanisms is the evidence that some key enzymes, besides their role in the maintenance of genome stability, display also unexpected noncanonical functions associated with RNA metabolism in specific subcellular districts (e.g., nucleoli). During the evolution of these key enzymes, the acquisition of unfolded domains significantly amplified the possibility to interact with different partners and substrates, possibly explaining their phylogenetic gain of functions.
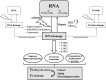
Recent advances: After nucleolar stress or DNA damage, many DNA repair proteins can freely relocalize from nucleoli to the nucleoplasm. This process may represent a surveillance mechanism to monitor the synthesis and correct assembly of ribosomal units affecting cell cycle progression or inducing p53-mediated apoptosis or senescence.
Critical issues: A paradigm for this kind of regulation is represented by some enzymes of the DNA base excision repair (BER) pathway, such as apurinic/apyrimidinic endonuclease 1 (APE1). In this review, the role of the nucleolus and the noncanonical functions of the APE1 protein are discussed in light of their possible implications in human pathologies.
Future directions: A productive cross-talk between DNA repair enzymes and proteins involved in RNA metabolism seems reasonable as the nucleolus is emerging as a dynamic functional hub that coordinates cell growth arrest and DNA repair mechanisms. These findings will drive further analyses on other BER proteins and might imply that nucleic acid processing enzymes are more versatile than originally thought having evolved DNA-targeted functions after a previous life in the early RNA world.
Figures



References
-
- Aas PA, Otterlei M, Falnes PO, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, and Krokan HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863, 2003 - PubMed
-
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, and Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11, 2002 - PubMed
-
- Andersen JS, Lam YW, Leung AKL, Ong S, Lyon CE, Lamond AI, and Mann M. Nucleolar proteome dynamics. Nature 433: 77–83, 2005 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

