A functional glycogen biosynthesis pathway in Lactobacillus acidophilus: expression and analysis of the glg operon
- PMID: 23879596
- PMCID: PMC4282360
- DOI: 10.1111/mmi.12338
A functional glycogen biosynthesis pathway in Lactobacillus acidophilus: expression and analysis of the glg operon
Abstract
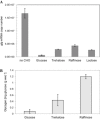
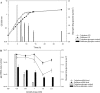
Glycogen metabolism contributes to energy storage and various physiological functions in some prokaryotes, including colonization persistence. A role for glycogen metabolism is proposed on the survival and fitness of Lactobacillus acidophilus, a probiotic microbe, in the human gastrointestinal environment. L. acidophilus NCFM possesses a glycogen metabolism (glg) operon consisting of glgBCDAP-amy-pgm genes. Expression of the glg operon and glycogen accumulation were carbon source- and growth phase-dependent, and were repressed by glucose. The highest intracellular glycogen content was observed in early log-phase cells grown on trehalose, which was followed by a drastic decrease of glycogen content prior to entering stationary phase. In raffinose-grown cells, however, glycogen accumulation gradually declined following early log phase and was maintained at stable levels throughout stationary phase. Raffinose also induced an overall higher temporal glg expression throughout growth compared with trehalose. Isogenic ΔglgA (glycogen synthase) and ΔglgB (glycogen-branching enzyme) mutants are glycogen-deficient and exhibited growth defects on raffinose. The latter observation suggests a reciprocal relationship between glycogen synthesis and raffinose metabolism. Deletion of glgB or glgP (glycogen phosphorylase) resulted in defective growth and increased bile sensitivity. The data indicate that glycogen metabolism is involved in growth maintenance, bile tolerance and complex carbohydrate utilization in L. acidophilus.
© 2013 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Figures




Similar articles
-
Insights into glycogen metabolism in Lactobacillus acidophilus: impact on carbohydrate metabolism, stress tolerance and gut retention.Microb Cell Fact. 2014 Nov 20;13:94. doi: 10.1186/s12934-014-0094-3. Microb Cell Fact. 2014. PMID: 25410006 Free PMC article.
-
Gene organization and transcription analysis of the Agrobacterium tumefaciens glycogen (glg) operon: two transcripts for the single phosphoglucomutase gene.J Bacteriol. 1998 Dec;180(24):6557-64. doi: 10.1128/JB.180.24.6557-6564.1998. J Bacteriol. 1998. PMID: 9851999 Free PMC article.
-
Analysis of Mesorhizobium loti glycogen operon: effect of phosphoglucomutase (pgm) and glycogen synthase (g/gA) null mutants on nodulation of Lotus tenuis.Mol Plant Microbe Interact. 2002 Apr;15(4):368-75. doi: 10.1094/MPMI.2002.15.4.368. Mol Plant Microbe Interact. 2002. PMID: 12026175
-
Glycogen branches out: new perspectives on the role of glycogen metabolism in the integration of metabolic pathways.Am J Physiol Endocrinol Metab. 2006 Jul;291(1):E1-8. doi: 10.1152/ajpendo.00652.2005. Epub 2006 Feb 14. Am J Physiol Endocrinol Metab. 2006. PMID: 16478770 Review.
-
New perspectives on the storage and organization of muscle glycogen.Can J Appl Physiol. 2002 Apr;27(2):179-203. doi: 10.1139/h02-012. Can J Appl Physiol. 2002. PMID: 12179957 Review.
Cited by
-
Identification of the Genes Related to the Glycogen Metabolism in Hyperthermophilic Archaeon, Sulfolobus acidocaldarius.Front Microbiol. 2021 May 13;12:661053. doi: 10.3389/fmicb.2021.661053. eCollection 2021. Front Microbiol. 2021. PMID: 34054761 Free PMC article.
-
Glycogen Metabolism in Candida albicans Impacts Fitness and Virulence during Vulvovaginal and Invasive Candidiasis.mBio. 2023 Apr 25;14(2):e0004623. doi: 10.1128/mbio.00046-23. Epub 2023 Feb 22. mBio. 2023. PMID: 36840583 Free PMC article.
-
Intracellular glycogen accumulation by human gut commensals as a niche adaptation trait.Gut Microbes. 2023 Jan-Dec;15(1):2235067. doi: 10.1080/19490976.2023.2235067. Gut Microbes. 2023. PMID: 37526383 Free PMC article. Review.
-
Insights into glycogen metabolism in Lactobacillus acidophilus: impact on carbohydrate metabolism, stress tolerance and gut retention.Microb Cell Fact. 2014 Nov 20;13:94. doi: 10.1186/s12934-014-0094-3. Microb Cell Fact. 2014. PMID: 25410006 Free PMC article.
-
Insight into Potential Probiotic Markers Predicted in Lactobacillus pentosus MP-10 Genome Sequence.Front Microbiol. 2017 May 22;8:891. doi: 10.3389/fmicb.2017.00891. eCollection 2017. Front Microbiol. 2017. PMID: 28588563 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

