Sub-inhibitory concentrations of some antibiotics can drive diversification of Pseudomonas aeruginosa populations in artificial sputum medium
- PMID: 23879797
- PMCID: PMC3726342
- DOI: 10.1186/1471-2180-13-170
Sub-inhibitory concentrations of some antibiotics can drive diversification of Pseudomonas aeruginosa populations in artificial sputum medium
Abstract
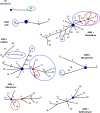
Background: Pseudomonas aeruginosa populations within the cystic fibrosis lung exhibit extensive phenotypic and genetic diversification. The resultant population diversity is thought to be crucial to the persistence of infection and may underpin the progression of disease. However, because cystic fibrosis lungs represent ecologically complex and hostile environments, the selective forces driving this diversification in vivo remain unclear. We took an experimental evolution approach to test the hypothesis that sub-inhibitory antibiotics can drive diversification of P. aeruginosa populations. Replicate populations of P. aeruginosa LESB58 were cultured for seven days in artificial sputum medium with and without sub-inhibitory concentrations of various clinically relevant antibiotics. We then characterised diversification with respect to 13 phenotypic and genotypic characteristics.
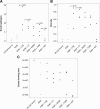
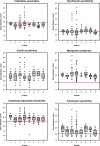
Results: We observed that higher population diversity evolved in the presence of azithromycin, ceftazidime or colistin relative to antibiotic-free controls. Divergence occurred due to alterations in antimicrobial susceptibility profiles following exposure to azithromycin, ceftazidime and colistin. Alterations in colony morphology and pyocyanin production were observed following exposure to ceftazidime and colistin only. Diversification was not observed in the presence of meropenem.
Conclusions: Our study indicates that certain antibiotics can promote population diversification when present in sub-inhibitory concentrations. Hence, the choice of antibiotic may have previously unforeseen implications for the development of P. aeruginosa infections in the lungs of cystic fibrosis patients.
Figures

Similar articles
-
Evolutionary diversification of Pseudomonas aeruginosa in an artificial sputum model.BMC Microbiol. 2017 Jan 5;17(1):3. doi: 10.1186/s12866-016-0916-z. BMC Microbiol. 2017. PMID: 28056789 Free PMC article.
-
Unveiling the early events of Pseudomonas aeruginosa adaptation in cystic fibrosis airway environment using a long-term in vitro maintenance.Int J Med Microbiol. 2018 Dec;308(8):1053-1064. doi: 10.1016/j.ijmm.2018.10.003. Epub 2018 Oct 11. Int J Med Microbiol. 2018. PMID: 30377031
-
Recombination is a key driver of genomic and phenotypic diversity in a Pseudomonas aeruginosa population during cystic fibrosis infection.Sci Rep. 2015 Jan 12;5:7649. doi: 10.1038/srep07649. Sci Rep. 2015. PMID: 25578031 Free PMC article.
-
Standard versus biofilm antimicrobial susceptibility testing to guide antibiotic therapy in cystic fibrosis.Cochrane Database Syst Rev. 2020 Jun 10;6(6):CD009528. doi: 10.1002/14651858.CD009528.pub5. Cochrane Database Syst Rev. 2020. PMID: 32520436 Free PMC article.
-
Diversification of Pseudomonas aeruginosa within the cystic fibrosis lung and its effects on antibiotic resistance.FEMS Microbiol Lett. 2018 Mar 1;365(6). doi: 10.1093/femsle/fny026. FEMS Microbiol Lett. 2018. PMID: 29401362 Review.
Cited by
-
Genomics of Diversification of Pseudomonas aeruginosa in Cystic Fibrosis Lung-like Conditions.Genome Biol Evol. 2022 May 31;14(6):evac074. doi: 10.1093/gbe/evac074. Genome Biol Evol. 2022. PMID: 35660861 Free PMC article.
-
Maximum levels of cross-contamination for 24 antimicrobial active substances in non-target feed. Part 9: Polymyxins: colistin.EFSA J. 2021 Oct 26;19(10):e06861. doi: 10.2903/j.efsa.2021.6861. eCollection 2021 Oct. EFSA J. 2021. PMID: 34729089 Free PMC article.
-
The Effects of Sub-inhibitory Antibiotic Concentrations on Pseudomonas aeruginosa: Reduced Susceptibility Due to Mutations.Front Microbiol. 2021 Dec 20;12:789550. doi: 10.3389/fmicb.2021.789550. eCollection 2021. Front Microbiol. 2021. PMID: 34987489 Free PMC article.
-
Ecological and evolutionary mechanisms driving within-patient emergence of antimicrobial resistance.Nat Rev Microbiol. 2024 Oct;22(10):650-665. doi: 10.1038/s41579-024-01041-1. Epub 2024 Apr 30. Nat Rev Microbiol. 2024. PMID: 38689039 Review.
-
TpiA is a Key Metabolic Enzyme That Affects Virulence and Resistance to Aminoglycoside Antibiotics through CrcZ in Pseudomonas aeruginosa.mBio. 2020 Jan 7;11(1):e02079-19. doi: 10.1128/mBio.02079-19. mBio. 2020. PMID: 31911486 Free PMC article.
References
-
- Teichgraber V, Ulrich M, Endlich N, Riethmuller J, Wilker B, De Oliveira-Munding CC, van Heeckeren AM, Barr ML, von Kürthy G, Schmid KW, Weller M, Tümmler B, Lang F, Grassme H, Döring G, Gulbins E. Ceramide accumulation mediates inflammation, cell death and infection susceptibility in cystic fibrosis. Nat Med. 2008;14:382–391. doi: 10.1038/nm1748. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

