Genetic regulation of cold-induced albinism in the maize inbred line A661
- PMID: 23881393
- PMCID: PMC3745721
- DOI: 10.1093/jxb/ert189
Genetic regulation of cold-induced albinism in the maize inbred line A661
Abstract
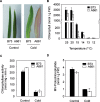
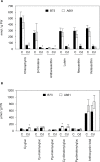
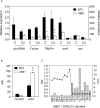
In spite of multiple studies elucidating the regulatory pathways controlling chlorophyll biosynthesis and photosynthetic activity, little is known about the molecular mechanism regulating cold-induced chlorosis in higher plants. Herein the characterization of the maize inbred line A661 which shows a cold-induced albino phenotype is reported. The data show that exposure of seedlings to low temperatures during early leaf biogenesis led to chlorophyll losses in this inbred. A661 shows a high plasticity, recovering resting levels of photosynthesis activity when exposed to optimal temperatures. Biochemical and transcriptome data indicate that at suboptimal temperatures chlorophyll could not be fully accommodated in the photosynthetic antenna in A661, remaining free in the chloroplast. The accumulation of free chlorophyll activates the expression of an early light inducible protein (elip) gene which binds chlorophyll to avoid cross-reactions that could lead to the generation of harmful reactive oxygen species. Higher levels of the elip transcript were observed in plants showing a cold-induced albino phenotype. Forward genetic analysis reveals that a gene located on the short arm of chromosome 2 regulates this protective mechanism.
Keywords: Albinism; Zea mays.; chlorophyll biosynthesis; cold; early light inducible protein (elip); photosystem.
Figures

References
-
- Aarti D, Tanaka R, Ito H, Tanaka A. 2007. High light inhibits chlorophyll biosynthesis at the level of 5-aminolevulinate synthesis during de-etiolation in cucumber (Cucumis sativus) cotyledons. Photochemistry and Photobiology 83, 171–176. - PubMed
-
- Adamska I, Kloppstech K, Ohad I. 1993. Early light-inducible protein in pea is stable during light stress but is degraded during recovery at low light intensity. Journal of Biological Chemistry 268, 5438–5444. - PubMed
-
- Agne B, Kessler F. 2009. Protein transport in organelles: the Toc complex way of preprotein import. FEBS Journal 276, 1156–1165. - PubMed
-
- Beale SI. 1999. Enzymes of chlorophyll biosynthesis. Photosynthesis Research 60, 43–73.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

