Helicase activation and establishment of replication forks at chromosomal origins of replication
- PMID: 23881938
- PMCID: PMC3839609
- DOI: 10.1101/cshperspect.a010371
Helicase activation and establishment of replication forks at chromosomal origins of replication
Abstract
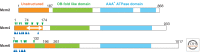
Many replication proteins assemble on the pre-RC-formed replication origins and constitute the pre-initiation complex (pre-IC). This complex formation facilitates the conversion of Mcm2-7 in the pre-RC to an active DNA helicase, the Cdc45-Mcm-GINS (CMG) complex. Two protein kinases, cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK), work to complete the formation of the pre-IC. Each kinase is responsible for a distinct step of the process in yeast; Cdc45 associates with origins in a DDK-dependent manner, whereas the association of GINS with origins depends on CDK. These associations with origins also require specific initiation proteins: Sld3 for Cdc45; and Dpb11, Sld2, and Sld3 for GINS. Functional homologs of these proteins exist in metazoa, although pre-IC formation cannot be separated by requirement of DDK and CDK because of experimental limitations. Once the replicative helicase is activated, the origin DNA is unwound, and bidirectional replication forks are established.
Figures


References
-
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3: 958–965 - PubMed
-
- Arias EE, Walter JC 2007. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 - PubMed
-
- Bachrati CZ, Hickson ID 2008. RecQ helicases: Guardian angels of the DNA replication fork. Chromosoma 117: 219–233 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
