Break-induced DNA replication
- PMID: 23881940
- PMCID: PMC3839615
- DOI: 10.1101/cshperspect.a010397
Break-induced DNA replication
Abstract
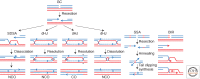
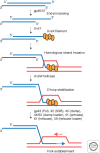
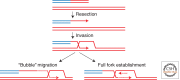
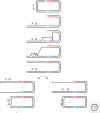
Recombination-dependent DNA replication, often called break-induced replication (BIR), was initially invoked to explain recombination events in bacteriophage but it has recently been recognized as a fundamentally important mechanism to repair double-strand chromosome breaks in eukaryotes. This mechanism appears to be critically important in the restarting of stalled and broken replication forks and in maintaining the integrity of eroded telomeres. Although BIR helps preserve genome integrity during replication, it also promotes genome instability by the production of loss of heterozygosity and the formation of nonreciprocal translocations, as well as in the generation of complex chromosomal rearrangements.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM076020/GM/NIGMS NIH HHS/United States
- GM20056/GM/NIGMS NIH HHS/United States
- R01 GM051753/GM/NIGMS NIH HHS/United States
- F32 GM096690/GM/NIGMS NIH HHS/United States
- R01 GM061766/GM/NIGMS NIH HHS/United States
- R37 GM020056/GM/NIGMS NIH HHS/United States
- T32 GM007122/GM/NIGMS NIH HHS/United States
- GM51753/GM/NIGMS NIH HHS/United States
- 2T32GM007122/GM/NIGMS NIH HHS/United States
- GM76020/GM/NIGMS NIH HHS/United States
- GM61766/GM/NIGMS NIH HHS/United States
- R01 GM020056/GM/NIGMS NIH HHS/United States
- 1F32GM096690-01/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
