Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids
- PMID: 23882013
- PMCID: PMC3735187
- DOI: 10.1128/mBio.00281-13
Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids
Abstract
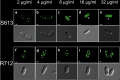
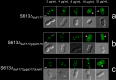
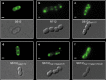
Treatment of multidrug-resistant enterococci has become a challenging clinical problem in hospitals around the world due to the lack of reliable therapeutic options. Daptomycin (DAP), a cell membrane-targeting cationic antimicrobial lipopeptide, is the only antibiotic with in vitro bactericidal activity against vancomycin-resistant enterococci (VRE). However, the clinical use of DAP against VRE is threatened by emergence of resistance during therapy, but the mechanisms leading to DAP resistance are not fully understood. The mechanism of action of DAP involves interactions with the cell membrane in a calcium-dependent manner, mainly at the level of the bacterial septum. Previously, we demonstrated that development of DAP resistance in vancomycin-resistant Enterococcus faecalis is associated with mutations in genes encoding proteins with two main functions, (i) control of the cell envelope stress response to antibiotics and antimicrobial peptides (LiaFSR system) and (ii) cell membrane phospholipid metabolism (glycerophosphoryl diester phosphodiesterase and cardiolipin synthase). In this work, we show that these VRE can resist DAP-elicited cell membrane damage by diverting the antibiotic away from its principal target (division septum) to other distinct cell membrane regions. DAP septal diversion by DAP-resistant E. faecalis is mediated by initial redistribution of cell membrane cardiolipin-rich microdomains associated with a single amino acid deletion within the transmembrane protein LiaF (a member of a three-component regulatory system [LiaFSR] involved in cell envelope homeostasis). Full expression of DAP resistance requires additional mutations in enzymes (glycerophosphoryl diester phosphodiesterase and cardiolipin synthase) that alter cell membrane phospholipid content. Our findings describe a novel mechanism of bacterial resistance to cationic antimicrobial peptides.
Importance: The emergence of antibiotic resistance in bacterial pathogens is a threat to public health. Understanding the mechanisms of resistance is of crucial importance to develop new strategies to combat multidrug-resistant microorganisms. Vancomycin-resistant enterococci (VRE) are one of the most recalcitrant hospital-associated pathogens against which new therapies are urgently needed. Daptomycin (DAP) is a calcium-decorated antimicrobial lipopeptide whose target is the bacterial cell membrane. A current paradigm suggests that Gram-positive bacteria become resistant to cationic antimicrobial peptides via an electrostatic repulsion of the antibiotic molecule from a more positively charged cell surface. In this work, we provide evidence that VRE use a novel strategy to avoid DAP-elicited killing. Instead of "repelling" the antibiotic from the cell surface, VRE diverts the antibiotic molecule from the septum and "traps" it in distinct membrane regions. We provide genetic and biochemical bases responsible for the mechanism of resistance and disclose new targets for potential antimicrobial development.
Figures
Similar articles
-
Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content.PLoS One. 2012;7(8):e43958. doi: 10.1371/journal.pone.0043958. Epub 2012 Aug 27. PLoS One. 2012. PMID: 22952824 Free PMC article.
-
Molecular basis of cell membrane adaptation in daptomycin-resistant Enterococcus faecalis.JCI Insight. 2024 Nov 22;9(22):e173836. doi: 10.1172/jci.insight.173836. JCI Insight. 2024. PMID: 39405116 Free PMC article.
-
A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis.J Infect Dis. 2015 Apr 15;211(8):1317-25. doi: 10.1093/infdis/jiu602. Epub 2014 Oct 31. J Infect Dis. 2015. PMID: 25362197 Free PMC article.
-
Mechanisms of drug resistance: daptomycin resistance.Ann N Y Acad Sci. 2015 Sep;1354:32-53. doi: 10.1111/nyas.12948. Epub 2015 Oct 23. Ann N Y Acad Sci. 2015. PMID: 26495887 Free PMC article. Review.
-
Mechanism of Action and Resistance to Daptomycin in Staphylococcus aureus and Enterococci.Cold Spring Harb Perspect Med. 2016 Nov 1;6(11):a026997. doi: 10.1101/cshperspect.a026997. Cold Spring Harb Perspect Med. 2016. PMID: 27580748 Free PMC article. Review.
Cited by
-
The lipid-modifying multiple peptide resistance factor is an oligomer consisting of distinct interacting synthase and flippase subunits.mBio. 2015 Jan 27;6(1):e02340-14. doi: 10.1128/mBio.02340-14. mBio. 2015. PMID: 25626904 Free PMC article.
-
Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer.Antimicrob Agents Chemother. 2015 Jul;59(7):4094-105. doi: 10.1128/AAC.00344-15. Epub 2015 Apr 27. Antimicrob Agents Chemother. 2015. PMID: 25918141 Free PMC article.
-
Bacteriophage-Antibiotic Combinations for Enterococcus faecium with Varying Bacteriophage and Daptomycin Susceptibilities.Antimicrob Agents Chemother. 2020 Aug 20;64(9):e00993-20. doi: 10.1128/AAC.00993-20. Print 2020 Aug 20. Antimicrob Agents Chemother. 2020. PMID: 32571816 Free PMC article.
-
Polymyxin and lipopeptide antibiotics: membrane-targeting drugs of last resort.Microbiology (Reading). 2022 Feb;168(2):001136. doi: 10.1099/mic.0.001136. Microbiology (Reading). 2022. PMID: 35118938 Free PMC article. Review.
-
Evolution of Enterococcus faecium in Response to a Combination of Daptomycin and Fosfomycin Reveals Distinct and Diverse Adaptive Strategies.Antimicrob Agents Chemother. 2022 Jun 21;66(6):e0233321. doi: 10.1128/aac.02333-21. Epub 2022 May 11. Antimicrob Agents Chemother. 2022. PMID: 35543524 Free PMC article.
References
-
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America Clin. Infect. Dis. 48:1–12 - PubMed
-
- Munoz-Price LS, Lolans K, Quinn JP. 2005. Emergence of resistance to daptomycin during treatment of vancomycin-resistant Enterococcus faecalis infection. Clin. Infect. Dis. 41:565–566 - PubMed
-
- Muraih JK, Pearson A, Silverman J, Palmer M. 2011. Oligomerization of daptomycin on membranes. Biochim. Biophys. Acta 1808:1154–1160 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

