Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species
- PMID: 23882017
- PMCID: PMC3735181
- DOI: 10.1128/mBio.00480-13
Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species
Abstract
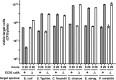
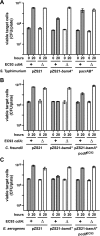
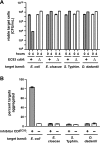
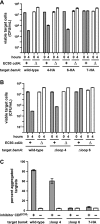
Bacteria that express contact-dependent growth inhibition (CDI) systems outcompete siblings that lack immunity, suggesting that CDI mediates intercellular competition. To further explore the role of CDI in competition, we determined the target cell range of the CDIEC93 system from Escherichia coli EC93. The CdiAEC93 effector protein recognizes the widely conserved BamA protein as a receptor, yet E. coli EC93 does not inhibit other enterobacterial species. The predicted membrane topology of BamA indicates that three of its extracellular loops vary considerably between species, suggesting that loop heterogeneity may control CDI specificity. Consistent with this hypothesis, other enterobacteria are sensitized to CDIEC93 upon the expression of E. coli bamA and E. coli cells become CDIEC93 resistant when bamA is replaced with alleles from other species. Our data indicate that BamA loops 6 and 7 form the CdiAEC93-binding epitope and their variation between species restricts CDIEC93 target cell selection. Although BamA loops 6 and 7 vary dramatically between species, these regions are identical in hundreds of E. coli strains, suggesting that BamAEcoli and CdiAEC93 play a role in self-nonself discrimination.
Importance: Contact-dependent growth inhibition (CDI) systems are widespread among Gram-negative bacteria, enabling them to bind to neighboring bacterial cells and deliver protein toxins that inhibit cell growth. In this study, we tested the role of CDI in interspecies competition using intestinal isolate Escherichia coli EC93 as an inhibitor cell model. Although E. coli EC93 inhibits different E. coli strains, other bacterial species from the intestine are completely resistant to CDI. We show that resistance is due to small variations in the CDI receptor that prevent other species from being recognized as target cells. CDI receptor interactions thus provide a mechanism by which bacteria can distinguish siblings and other close relatives (self) from more distant relatives or other species of bacteria (nonself). Our results provide a possible means by which antimicrobials could be directed to one or only a few related bacterial pathogens by using a specific receptor "zip code."
Figures


References
-
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245–1248 - PubMed
-
- Clantin B, Delattre AS, Rucktooa P, Saint N, Méli AC, Locht C, Jacob-Dubuisson F, Villeret V. 2007. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317:957–961 - PubMed
-
- Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC. 2001. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 42:279–292 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
