Network-based analysis of genome wide association data provides novel candidate genes for lipid and lipoprotein traits
- PMID: 23882023
- PMCID: PMC3820950
- DOI: 10.1074/mcp.M112.024851
Network-based analysis of genome wide association data provides novel candidate genes for lipid and lipoprotein traits
Abstract
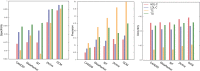
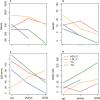
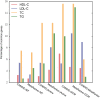
Genome wide association studies (GWAS) identify susceptibility loci for complex traits, but do not identify particular genes of interest. Integration of functional and network information may help in overcoming this limitation and identifying new susceptibility loci. Using GWAS and comorbidity data, we present a network-based approach to predict candidate genes for lipid and lipoprotein traits. We apply a prediction pipeline incorporating interactome, co-expression, and comorbidity data to Global Lipids Genetics Consortium (GLGC) GWAS for four traits of interest, identifying phenotypically coherent modules. These modules provide insights regarding gene involvement in complex phenotypes with multiple susceptibility alleles and low effect sizes. To experimentally test our predictions, we selected four candidate genes and genotyped representative SNPs in the Malmö Diet and Cancer Cardiovascular Cohort. We found significant associations with LDL-C and total-cholesterol levels for a synonymous SNP (rs234706) in the cystathionine beta-synthase (CBS) gene (p = 1 × 10(-5) and adjusted-p = 0.013, respectively). Further, liver samples taken from 206 patients revealed that patients with the minor allele of rs234706 had significant dysregulation of CBS (p = 0.04). Despite the known biological role of CBS in lipid metabolism, SNPs within the locus have not yet been identified in GWAS of lipoprotein traits. Thus, the GWAS-based Comorbidity Module (GCM) approach identifies candidate genes missed by GWAS studies, serving as a broadly applicable tool for the investigation of other complex disease phenotypes.
Conflict of interest statement
Conflict of interest: The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Genetic associations with expression for genes implicated in GWAS studies for atherosclerotic cardiovascular disease and blood phenotypes.Hum Mol Genet. 2014 Feb 1;23(3):782-95. doi: 10.1093/hmg/ddt461. Epub 2013 Sep 20. Hum Mol Genet. 2014. PMID: 24057673 Free PMC article.
-
Candidate gene analysis of the fibrinogen phenotype reveals the importance of polygenic co-regulation.Matrix Biol. 2017 Jul;60-61:16-26. doi: 10.1016/j.matbio.2016.10.005. Epub 2016 Oct 19. Matrix Biol. 2017. PMID: 27771416
-
Investigation of multi-trait associations using pathway-based analysis of GWAS summary statistics.BMC Genomics. 2019 Feb 4;20(Suppl 1):79. doi: 10.1186/s12864-018-5373-7. BMC Genomics. 2019. PMID: 30712509 Free PMC article.
-
The breadth and impact of the Global Lipids Genetics Consortium.Curr Opin Lipidol. 2025 Apr 1;36(2):61-70. doi: 10.1097/MOL.0000000000000966. Epub 2024 Dec 9. Curr Opin Lipidol. 2025. PMID: 39602359 Free PMC article. Review.
-
Integrated approaches to functionally characterize novel factors in lipoprotein metabolism.Curr Opin Lipidol. 2012 Apr;23(2):104-10. doi: 10.1097/MOL.0b013e328350fc3d. Curr Opin Lipidol. 2012. PMID: 22327609 Review.
Cited by
-
Exploring the cross-phenotype network region of disease modules reveals concordant and discordant pathways between chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis.Hum Mol Genet. 2019 Jul 15;28(14):2352-2364. doi: 10.1093/hmg/ddz069. Hum Mol Genet. 2019. PMID: 30997486 Free PMC article.
-
Using network clustering to predict copy number variations associated with health disparities.PeerJ. 2015 Mar 5;3:e677. doi: 10.7717/peerj.677. eCollection 2015. PeerJ. 2015. PMID: 25780754 Free PMC article.
-
Identifying genes targeted by disease-associated non-coding SNPs with a protein knowledge graph.PLoS One. 2022 Jul 13;17(7):e0271395. doi: 10.1371/journal.pone.0271395. eCollection 2022. PLoS One. 2022. PMID: 35830458 Free PMC article.
-
Next-generation gene discovery for variants of large impact on lipid traits.Curr Opin Lipidol. 2015 Apr;26(2):114-9. doi: 10.1097/MOL.0000000000000156. Curr Opin Lipidol. 2015. PMID: 25636063 Free PMC article. Review.
-
Network Medicine in the Age of Biomedical Big Data.Front Genet. 2019 Apr 11;10:294. doi: 10.3389/fgene.2019.00294. eCollection 2019. Front Genet. 2019. PMID: 31031797 Free PMC article. Review.
References
-
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., Hunter D. J., McCarthy M. I., Ramos E. M., Cardon L. R., Chakravarti A., Cho J. H., Guttmacher A. E., Kong A., Kruglyak L., Mardis E., Rotimi C. N., Slatkin M., Valle D., Whittemore A. S., Boehnke M., Clark A. G., Eichler E. E., Gibson G., Haines J. L., Mackay T. F., McCarroll S. A., Visscher P. M. (2009) Finding the missing heritability of complex diseases. Nature 461, 747–753 - PMC - PubMed
-
- Hegele R. A. (2010) Genome-wide association studies of plasma lipids: have we reached the limit? Arterioscler. Thromb. Vasc. Biol. 30, 2084–2086 - PubMed
-
- Wang K., Zhang H., Kugathasan S., Annese V., Bradfield J. P., Russell R. K., Sleiman P. M., Imielinski M., Glessner J., Hou C., Wilson D. C., Walters T., Kim C., Frackelton E. C., Lionetti P., Barabino A., Van Limbergen J., Guthery S., Denson L., Piccoli D., Li M., Dubinsky M., Silverberg M., Griffiths A., Grant S. F., Satsangi J., Baldassano R., Hakonarson H. (2009) Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am. J. Hum. Genet. 84, 399–405 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

