The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal
- PMID: 23882114
- PMCID: PMC3753931
- DOI: 10.1261/rna.039255.113
The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal
Abstract
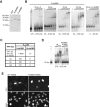
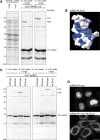
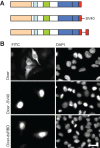
Dicer is a key player in microRNA (miRNA) and RNA interference (RNAi) pathways, processing miRNA precursors and double-stranded RNA into ∼21-nt-long products ultimately triggering sequence-dependent gene silencing. Although processing of substrates in vertebrate cells occurs in the cytoplasm, there is growing evidence suggesting Dicer is also present and functional in the nucleus. To address this possibility, we searched for a nuclear localization signal (NLS) in human Dicer and identified its C-terminal double-stranded RNA binding domain (dsRBD) as harboring NLS activity. We show that the dsRBD-NLS can mediate nuclear import of a reporter protein via interaction with importins β, 7, and 8. In the context of full-length Dicer, the dsRBD-NLS is masked. However, duplication of the dsRBD localizes the full-length protein to the nucleus. Furthermore, deletion of the N-terminal helicase domain results in partial accumulation of Dicer in the nucleus upon leptomycin B treatment, indicating that CRM1 contributes to nuclear export of Dicer. Finally, we demonstrate that human Dicer has the ability to shuttle between the nucleus and the cytoplasm. We conclude that Dicer is a shuttling protein whose steady-state localization is cytoplasmic.
Keywords: Dicer; NLS; RNAi; dsRBD; helicase.
Figures





Similar articles
-
Cryo-EM structures of human DICER dicing a pre-miRNA substrate.FEBS J. 2024 Jul;291(14):3072-3079. doi: 10.1111/febs.17048. Epub 2024 Jan 10. FEBS J. 2024. PMID: 38151772 Review.
-
The role of human Dicer-dsRBD in processing small regulatory RNAs.PLoS One. 2012;7(12):e51829. doi: 10.1371/journal.pone.0051829. Epub 2012 Dec 13. PLoS One. 2012. PMID: 23272173 Free PMC article.
-
A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1.Proc Natl Acad Sci U S A. 2014 May 6;111(18):E1852-61. doi: 10.1073/pnas.1323698111. Epub 2014 Apr 21. Proc Natl Acad Sci U S A. 2014. PMID: 24753571 Free PMC article.
-
Functions of double-stranded RNA-binding domains in nucleocytoplasmic transport.RNA Biol. 2014;11(10):1226-32. doi: 10.4161/15476286.2014.972856. RNA Biol. 2014. PMID: 25584639 Free PMC article. Review.
-
Nuclear phosphorylated Dicer processes double-stranded RNA in response to DNA damage.J Cell Biol. 2017 Aug 7;216(8):2373-2389. doi: 10.1083/jcb.201612131. Epub 2017 Jun 22. J Cell Biol. 2017. PMID: 28642363 Free PMC article.
Cited by
-
The Cajal body and the nucleolus: "In a relationship" or "It's complicated"?RNA Biol. 2017 Jun 3;14(6):739-751. doi: 10.1080/15476286.2016.1236169. Epub 2016 Sep 23. RNA Biol. 2017. PMID: 27661468 Free PMC article. Review.
-
The role of Dicer1 in the male reproductive tract.Asian J Androl. 2015 Sep-Oct;17(5):737-41. doi: 10.4103/1008-682X.155542. Asian J Androl. 2015. PMID: 25994652 Free PMC article. Review.
-
Endogenous RNA interference is driven by copy number.Elife. 2014 Feb 11;3:e01581. doi: 10.7554/eLife.01581. Elife. 2014. PMID: 24520161 Free PMC article.
-
RNA-binding proteins in pluripotency, differentiation, and reprogramming.Front Biol (Beijing). 2014 Oct;9(5):389-409. doi: 10.1007/s11515-014-1326-y. Front Biol (Beijing). 2014. PMID: 25554730 Free PMC article.
-
The Role of MicroRNAs in Liver Functioning: from Biogenesis to Therapeutic Approaches (Review).Sovrem Tekhnologii Med. 2023;15(5):54-79. doi: 10.17691/stm2023.15.5.06. Epub 2023 Oct 30. Sovrem Tekhnologii Med. 2023. PMID: 39967915 Free PMC article. Review.
References
-
- Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, et al. 2008. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 15: 268–279 - PMC - PubMed
-
- Blaszczyk J, Gan J, Tropea JE, Court DL, Waugh DS, Ji X 2004. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure 12: 457–466 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
