Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B
- PMID: 23884607
- PMCID: PMC3744726
- DOI: 10.1101/gad.220095.113
Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B
Abstract
The conversion of male germ cell chromatin to a nucleoprotamine structure is fundamental to the life cycle, yet the underlying molecular details remain obscure. Here we show that an essential step is the genome-wide incorporation of TH2B, a histone H2B variant of hitherto unknown function. Using mouse models in which TH2B is depleted or C-terminally modified, we show that TH2B directs the final transformation of dissociating nucleosomes into protamine-packed structures. Depletion of TH2B induces compensatory mechanisms that permit histone removal by up-regulating H2B and programming nucleosome instability through targeted histone modifications, including lysine crotonylation and arginine methylation. Furthermore, after fertilization, TH2B reassembles onto the male genome during protamine-to-histone exchange. Thus, TH2B is a unique histone variant that plays a key role in the histone-to-protamine packing of the male genome and guides genome-wide chromatin transitions that both precede and follow transmission of the male genome to the egg.
Keywords: BRDT; H2AZ; histone eviction; male contraception; male infertility; reprogramming; sex chromosome inactivation.
Figures
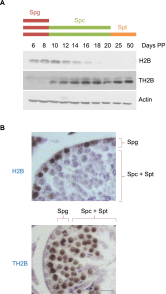




Comment in
-
How mammals pack their sperm: a variant matter.Genes Dev. 2013 Aug 1;27(15):1635-9. doi: 10.1101/gad.226167.113. Genes Dev. 2013. PMID: 23913918 Free PMC article.
-
Histone variants and sensing of chromatin functional states.Nucleus. 2013 Nov-Dec;4(6):438-42. doi: 10.4161/nucl.27088. Epub 2013 Nov 8. Nucleus. 2013. PMID: 24213375 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
