Construction of a Sonchus Yellow Net Virus minireplicon: a step toward reverse genetic analysis of plant negative-strand RNA viruses
- PMID: 23885070
- PMCID: PMC3807423
- DOI: 10.1128/JVI.01397-13
Construction of a Sonchus Yellow Net Virus minireplicon: a step toward reverse genetic analysis of plant negative-strand RNA viruses
Erratum in
- J Virol. 2013 Dec;87(23):13081
Abstract
Reverse genetic analyses of negative-strand RNA (NSR) viruses have provided enormous advances in our understanding of animal viruses over the past 20 years, but technical difficulties have hampered application to plant NSR viruses. To develop a reverse genetic approach for analysis of plant NSR viruses, we have engineered Sonchus yellow net nucleorhabdovirus (SYNV) minireplicon (MR) reporter cassettes for Agrobacterium tumefaciens expression in Nicotiana benthamiana leaves. Fluorescent reporter genes substituted for the SYNV N and P protein open reading frames (ORFs) exhibited intense single-cell foci throughout regions of infiltrated leaves expressing the SYNV MR derivatives and the SYNV nucleocapsid (N), phosphoprotein (P), and polymerase (L) proteins. Genomic RNA and mRNA transcription was detected for reporter genes substituted for both the SYNV N and P ORFs. These activities required expression of the N, P, and L core proteins in trans and were enhanced by codelivery of viral suppressor proteins that interfere with host RNA silencing. As is the case with other members of the Mononegavirales, we detected polar expression of fluorescent proteins and chloramphenicol acetyltransferase substitutions for the N and P protein ORFs. We also demonstrated the utility of the SYNV MR system for functional analysis of SYNV core proteins in trans and the cis-acting leader and trailer sequence requirements for transcription and replication. This work provides a platform for construction of more complex SYNV reverse genetic derivatives and presents a general strategy for reverse genetic applications with other plant NSR viruses.
Figures

 , leader sequence; N5′UTR, 5′ untranslated region of N protein mRNA; NPJ/P5′UTR, gene junction separating the N and P protein ORFs and the 5′ untranslated region of the P protein mRNA; L3′UTR, 3′ untranslated region of the L protein mRNA;
, leader sequence; N5′UTR, 5′ untranslated region of N protein mRNA; NPJ/P5′UTR, gene junction separating the N and P protein ORFs and the 5′ untranslated region of the P protein mRNA; L3′UTR, 3′ untranslated region of the L protein mRNA;  , SYNV trailer sequence; ΔRz = hepatitis delta virus ribozyme; NOS, pGD plasmid synthetase terminator. (B) Comparisons of GFP expression from the pSYNV-MRrsGFP-CAT minireplicon and pGD-GFP. Top panels, appearance of punctate GFP foci in N. benthamiana leaf tissue monitored with a Zeiss Lumar epifluorescence microscope for 14 days postinfiltration (dpi) with bacteria harboring the pSYNV-MReGFP-DsRed reporter cassette and the SYNV N, P, and L plasmids. The infiltration mixtures also contained bacteria with plasmids encoding the p19 and γb host gene silencing suppressors. The magnification bar represents 100 μm. GFP fluorescence from pSYNV-MReGFP-DsRed was evident at 4 to 6 dpi, and the foci continued to express GFP for up to 3 weeks postinfiltration until the infiltrated tissue began to senesce (not shown). Middle panels, micrographs at 10 dpi, showing punctate GFP foci in single cells of N. benthamiana leaf tissue expressing the pSYNV-MRrsGFP-CAT reporter cassette. Aside from the pSYNV-MRrsGFP-CAT reporter cassette, the infiltration mixtures were as in the top panels. Different magnifications are shown to illustrate the distribution of the foci and to demonstrate that the foci are confined to single cells. The magnification bar represents 100 μm. Bottom panels, fluorescence from leaf tissue infiltrated with pGD-GFP. Fluorescence was evident by 3 dpi and was evenly distributed throughout infiltrated zones rather than in punctate foci. Fluorescence was most intense at 6 dpi and declined in the 9- and 14-dpi periods. (C) TLC analysis of CAT activity. CAT activities from leaf extracts were analyzed by thin-layer chromatography (TLC) as described in Materials and Methods. M indicates marker chloramphenicol derivatives separated by TLC. CK denotes control leaf tissue infiltrated with pSYNV-MRrsGFP-CAT alone; 5 dpi and 10 dpi show CAT activity in N. benthamiana leaves harvested at 5 and 10 dpi. Positions of BCAM (nonacetylated fluorescent chloramphenicol substrate) and the CAT products 1Ac-BCAM (fluorescent 1-monoacetylated chloramphenicol), 3Ac-BCAM (fluorescent 3-monoacetylated chloramphenicol), and 1,3Ac-BCAM (fluorescent 1,3-diacetylated chloramphenicol) are indicated.
, SYNV trailer sequence; ΔRz = hepatitis delta virus ribozyme; NOS, pGD plasmid synthetase terminator. (B) Comparisons of GFP expression from the pSYNV-MRrsGFP-CAT minireplicon and pGD-GFP. Top panels, appearance of punctate GFP foci in N. benthamiana leaf tissue monitored with a Zeiss Lumar epifluorescence microscope for 14 days postinfiltration (dpi) with bacteria harboring the pSYNV-MReGFP-DsRed reporter cassette and the SYNV N, P, and L plasmids. The infiltration mixtures also contained bacteria with plasmids encoding the p19 and γb host gene silencing suppressors. The magnification bar represents 100 μm. GFP fluorescence from pSYNV-MReGFP-DsRed was evident at 4 to 6 dpi, and the foci continued to express GFP for up to 3 weeks postinfiltration until the infiltrated tissue began to senesce (not shown). Middle panels, micrographs at 10 dpi, showing punctate GFP foci in single cells of N. benthamiana leaf tissue expressing the pSYNV-MRrsGFP-CAT reporter cassette. Aside from the pSYNV-MRrsGFP-CAT reporter cassette, the infiltration mixtures were as in the top panels. Different magnifications are shown to illustrate the distribution of the foci and to demonstrate that the foci are confined to single cells. The magnification bar represents 100 μm. Bottom panels, fluorescence from leaf tissue infiltrated with pGD-GFP. Fluorescence was evident by 3 dpi and was evenly distributed throughout infiltrated zones rather than in punctate foci. Fluorescence was most intense at 6 dpi and declined in the 9- and 14-dpi periods. (C) TLC analysis of CAT activity. CAT activities from leaf extracts were analyzed by thin-layer chromatography (TLC) as described in Materials and Methods. M indicates marker chloramphenicol derivatives separated by TLC. CK denotes control leaf tissue infiltrated with pSYNV-MRrsGFP-CAT alone; 5 dpi and 10 dpi show CAT activity in N. benthamiana leaves harvested at 5 and 10 dpi. Positions of BCAM (nonacetylated fluorescent chloramphenicol substrate) and the CAT products 1Ac-BCAM (fluorescent 1-monoacetylated chloramphenicol), 3Ac-BCAM (fluorescent 3-monoacetylated chloramphenicol), and 1,3Ac-BCAM (fluorescent 1,3-diacetylated chloramphenicol) are indicated.

 ) probe. (C) Hybridization of genomic-sense RNAs in N. benthamiana extracts to a positive-sense trailer (
) probe. (C) Hybridization of genomic-sense RNAs in N. benthamiana extracts to a positive-sense trailer ( ) probe complementary to the 3′ end of the gRNAs. (D) Antigenomic RNAs hybridizing to a negative-sense rsGFP probe. A hybridizing RNA appearing slightly above the 720-nt GFP ORF size marker (GFP mRNA) was observed only in N. benthamiana leaves infiltrated with a complete mixture of plasmids expressing pSYNV-MRrsGFP-CAT and the SYNV N, P, and L proteins. The location of a marker corresponding in size to the 1,791-nt “positive-sense” (antigenomic) RNA is shown along the side of the panel. Only minor amounts of a hybridizing band corresponding to this RNA were detected in any of the hybridizations, and these are variable, suggesting that progeny agRNA transcripts are not amplified. (E) Detection of SYNV-MR agRNAs hybridizing to the negative-sense CAT probe. A 660-nt hybridizing band, corresponding to the CAT mRNA, hybridized to the probe only in tissue infiltrated with derivatives harboring pSYNV-MR and plasmids for expression of the N, P, and L proteins. (F) Analysis of SYNV agRNAs hybridizing to a negative-sense leader (
) probe complementary to the 3′ end of the gRNAs. (D) Antigenomic RNAs hybridizing to a negative-sense rsGFP probe. A hybridizing RNA appearing slightly above the 720-nt GFP ORF size marker (GFP mRNA) was observed only in N. benthamiana leaves infiltrated with a complete mixture of plasmids expressing pSYNV-MRrsGFP-CAT and the SYNV N, P, and L proteins. The location of a marker corresponding in size to the 1,791-nt “positive-sense” (antigenomic) RNA is shown along the side of the panel. Only minor amounts of a hybridizing band corresponding to this RNA were detected in any of the hybridizations, and these are variable, suggesting that progeny agRNA transcripts are not amplified. (E) Detection of SYNV-MR agRNAs hybridizing to the negative-sense CAT probe. A 660-nt hybridizing band, corresponding to the CAT mRNA, hybridized to the probe only in tissue infiltrated with derivatives harboring pSYNV-MR and plasmids for expression of the N, P, and L proteins. (F) Analysis of SYNV agRNAs hybridizing to a negative-sense leader ( ) probe. The intense hybridization corresponding to the 144-nt
) probe. The intense hybridization corresponding to the 144-nt  RNA was present only in tissue infiltrated with derivatives harboring the pSYNV MR and plasmids for expression of the core proteins.
RNA was present only in tissue infiltrated with derivatives harboring the pSYNV MR and plasmids for expression of the core proteins.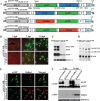

 , 18-nt deletion at the 5′ terminus of the
, 18-nt deletion at the 5′ terminus of the  sequence of pSYNV-MReGFP-DsRed; Δ
sequence of pSYNV-MReGFP-DsRed; Δ , 18-nt deletion at the 3′ terminus of pSYNV-MRGFP-DsRed; ΔHRz, HRz deletion preceding pSYNV-MReGFP-DsRed to prevent transcript processing. The lower panel depicts a Western blot probe of GFP accumulation in tissue infiltrated with the derivatives shown in the other panels.
, 18-nt deletion at the 3′ terminus of pSYNV-MRGFP-DsRed; ΔHRz, HRz deletion preceding pSYNV-MReGFP-DsRed to prevent transcript processing. The lower panel depicts a Western blot probe of GFP accumulation in tissue infiltrated with the derivatives shown in the other panels.References
-
- Pringle CR. 2005. Order Mononegavirales, p 609–614 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses Academic Press
-
- Tordo N, Benmansour A, Calisher C, Dietzgen RG, Fang RX, Jackson AO, Kurath G, Nadin-Davis S, Tesh RB, Walker PJ. 2005. Family Rhabdoviridae, p 626–644 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA
-
- Luytjes W, Krystal M, Enami M, Parvin JD, Palese P. 1989. Amplification, expression, and packaging of foreign gene by influenza virus. Cell 59:1107–1113 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

