Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment
- PMID: 23885072
- PMCID: PMC3807400
- DOI: 10.1128/JVI.01370-13
Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment
Abstract
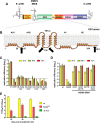
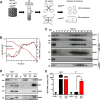
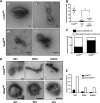
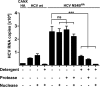
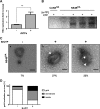
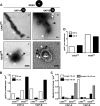
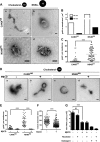
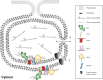
Like all other positive-strand RNA viruses, hepatitis C virus (HCV) induces rearrangements of intracellular membranes that are thought to serve as a scaffold for the assembly of the viral replicase machinery. The most prominent membranous structures present in HCV-infected cells are double-membrane vesicles (DMVs). However, their composition and role in the HCV replication cycle are poorly understood. To gain further insights into the biochemcial properties of HCV-induced membrane alterations, we generated a functional replicon containing a hemagglutinin (HA) affinity tag in nonstructural protein 4B (NS4B), the supposed scaffold protein of the viral replication complex. By using HA-specific affinity purification we isolated NS4B-containing membranes from stable replicon cells. Complementing biochemical and electron microscopy analyses of purified membranes revealed predominantly DMVs, which contained viral proteins NS3 and NS5A as well as enzymatically active viral replicase capable of de novo synthesis of HCV RNA. In addition to viral factors, co-opted cellular proteins, such as vesicle-associated membrane protein-associated protein A (VAP-A) and VAP-B, that are crucial for viral RNA replication, as well as cholesterol, a major structural lipid of detergent-resistant membranes, are highly enriched in DMVs. Here we describe the first isolation and biochemical characterization of HCV-induced DMVs. The results obtained underline their central role in the HCV replication cycle and suggest that DMVs are sites of viral RNA replication. The experimental approach described here is a powerful tool to more precisely define the molecular composition of membranous replication factories induced by other positive-strand RNA viruses, such as picorna-, arteri- and coronaviruses.
Figures

References
-
- Lavanchy D. 2009. The global burden of hepatitis C. Liver Int. 29(Suppl 1):74–81 - PubMed
-
- Sarrazin C, Hezode C, Zeuzem S, Pawlotsky JM. 2012. Antiviral strategies in hepatitis C virus infection. J. Hepatol. 56(Suppl 1):S88–S100 - PubMed
-
- Vermehren J, Susser S, Lange CM, Forestier N, Karey U, Hughes E, Ralston R, Tong X, Zeuzem S, Sarrazin C. 2012. Mutations selected in the hepatitis C virus NS3 protease domain during sequential treatment with boceprevir with and without pegylated interferon alfa-2b. J. Viral Hepat. 19:120–127 - PubMed
-
- van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB. (ed). 2000. Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA
-
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin I, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

