Circulating dendritic cells isolated from healthy seropositive donors are sites of human cytomegalovirus reactivation in vivo
- PMID: 23885077
- PMCID: PMC3807413
- DOI: 10.1128/JVI.01539-13
Circulating dendritic cells isolated from healthy seropositive donors are sites of human cytomegalovirus reactivation in vivo
Abstract
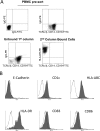
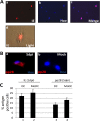
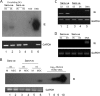
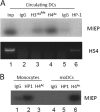
Primary infection with human cytomegalovirus (HCMV) is generally asymptomatic in healthy individuals and results in a lifelong infection of the host. In contrast, in immunosuppressed transplant recipients and late-stage AIDS patients, HCMV infection and reactivation can result in severe disease or death. In vivo, latency is established in bone marrow CD34(+) progenitor cells with reactivation linked with their differentiation to macrophages and dendritic cells (DCs). However, previous analyses have relied on ex vivo differentiation of myeloid progenitor cells to DCs in culture. Here, we now report on the isolation and analysis of circulating blood myeloid DCs, resulting from natural differentiation in vivo, from healthy HCMV-seropositive carriers. We show that these in vivo-differentiated circulating DCs are fully permissive for HCMV and exhibit a phenotype similar to that of monocyte-derived DCs routinely used for in vitro studies of HCMV. Importantly, we also show that these DCs from healthy HCMV-seropositive donors carry HCMV genomes and, significantly, are typically positive for viral immediate-early (IE) gene expression, in contrast to circulating monocytes, which carry genomes with an absence of IE expression. Finally, we show that HCMV reactivation from these circulating DCs is enhanced by inflammatory stimuli. Overall, these data argue that the differentiation in vivo of myeloid progenitors to circulating DCs promotes the reactivation of HCMV lytic gene expression in healthy individuals, thereby providing valuable confirmation of studies performed using in vitro generation of DCs from myeloid precursors to study HCMV reactivation.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

