Unraveling genomic variation from next generation sequencing data
- PMID: 23885890
- PMCID: PMC3726446
- DOI: 10.1186/1756-0381-6-13
Unraveling genomic variation from next generation sequencing data
Abstract
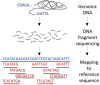
Elucidating the content of a DNA sequence is critical to deeper understand and decode the genetic information for any biological system. As next generation sequencing (NGS) techniques have become cheaper and more advanced in throughput over time, great innovations and breakthrough conclusions have been generated in various biological areas. Few of these areas, which get shaped by the new technological advances, involve evolution of species, microbial mapping, population genetics, genome-wide association studies (GWAs), comparative genomics, variant analysis, gene expression, gene regulation, epigenetics and personalized medicine. While NGS techniques stand as key players in modern biological research, the analysis and the interpretation of the vast amount of data that gets produced is a not an easy or a trivial task and still remains a great challenge in the field of bioinformatics. Therefore, efficient tools to cope with information overload, tackle the high complexity and provide meaningful visualizations to make the knowledge extraction easier are essential. In this article, we briefly refer to the sequencing methodologies and the available equipment to serve these analyses and we describe the data formats of the files which get produced by them. We conclude with a thorough review of tools developed to efficiently store, analyze and visualize such data with emphasis in structural variation analysis and comparative genomics. We finally comment on their functionality, strengths and weaknesses and we discuss how future applications could further develop in this field.
Figures








References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

