Design and generation of recombinant rabies virus vectors
- PMID: 23887178
- PMCID: PMC4028848
- DOI: 10.1038/nprot.2013.094
Design and generation of recombinant rabies virus vectors
Abstract
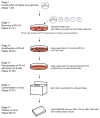
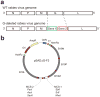
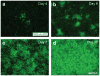
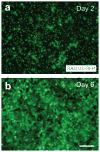
Rabies viruses, negative-strand RNA viruses, infect neurons through axon terminals and spread trans-synaptically in a retrograde direction between neurons. Rabies viruses whose glycoprotein (G) gene is deleted from the genome cannot spread across synapses. Complementation of G in trans, however, enables trans-synaptic spreading of G-deleted rabies viruses to directly connected, presynaptic neurons. Recombinant rabies viruses can encode genes of interest for labeling cells, controlling gene expression and monitoring or manipulating neural activity. Cre-dependent or bridge protein-mediated transduction and single-cell electroporation via the EnvA-TVA or EnvB-TVB (envelope glycoprotein and its specific receptor for avian sarcoma leukosis virus subgroup A or B) system allow cell type-specific or single cell-specific targeting. These rabies virus-based approaches permit the linking of connectivity to cell morphology and circuit function for particular cell types or single cells. Here we describe methods for construction of rabies viral vectors, recovery of G-deleted rabies viruses from cDNA, amplification of the viruses, pseudotyping them with EnvA or EnvB and concentration and titration of the viruses. The entire protocol takes 6-8 weeks.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

