Technical and implementation issues in using next-generation sequencing of cancers in clinical practice
- PMID: 23887607
- PMCID: PMC3749581
- DOI: 10.1038/bjc.2013.416
Technical and implementation issues in using next-generation sequencing of cancers in clinical practice
Abstract
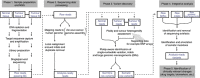
Next-generation sequencing (NGS) of cancer genomes promises to revolutionise oncology, with the ability to design and use targeted drugs, to predict outcome and response, and to classify tumours. It is continually becoming cheaper, faster and more reliable, with the capability to identify rare yet clinically important somatic mutations. Technical challenges include sequencing samples of low quality and/or quantity, reliable identification of structural and copy number variation, and assessment of intratumour heterogeneity. Once these problems are overcome, the use of the data to guide clinical decision making is not straightforward, and there is a risk of premature use of molecular changes to guide patient management in the absence of supporting evidence. Paradoxically, NGS may simply move the bottleneck of personalised medicine from data acquisition to the identification of reliable biomarkers. Standardised cancer NGS data collection on an international scale would be a significant step towards optimising patient care.
Figures
References
-
- Bao S, Jiang R, Kwan W. Evaluation of next generation sequencing software in mapping and assembly. J Hum Genet. 2011;56 (6:406–414. - PubMed
-
- Birol I, Jackman SD, Nielsen CB, Quian JQ, Varhol R, Stazyk G, Morin RD, Zhao Y, Hirst M, Schein JE, Hormans DE, Connors JM, Gascoyne RD, Marra MA, Jones SJ. De novo transcriptome assembly with ABySS. Bioinformatics. 2009;25 (21:2872–2877. - PubMed
-
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O'Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources