Long non-coding RNAs in haematological malignancies
- PMID: 23887658
- PMCID: PMC3759866
- DOI: 10.3390/ijms140815386
Long non-coding RNAs in haematological malignancies
Abstract
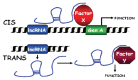
Long non-coding RNAs (lncRNAs) are functional RNAs longer than 200 nucleotides in length. LncRNAs are as diverse as mRNAs and they normally share the same biosynthetic machinery based on RNA polymerase II, splicing and polyadenylation. However, lncRNAs have low coding potential. Compared to mRNAs, lncRNAs are preferentially nuclear, more tissue specific and expressed at lower levels. Most of the lncRNAs described to date modulate the expression of specific genes by guiding chromatin remodelling factors; inducing chromosomal loopings; affecting transcription, splicing, translation or mRNA stability; or serving as scaffolds for the organization of cellular structures. They can function in cis, cotranscriptionally, or in trans, acting as decoys, scaffolds or guides. These functions seem essential to allow cell differentiation and growth. In fact, many lncRNAs have been shown to exert oncogenic or tumor suppressor properties in several cancers including haematological malignancies. In this review, we summarize what is known about lncRNAs, the mechanisms for their regulation in cancer and their role in leukemogenesis, lymphomagenesis and hematopoiesis. Furthermore, we discuss the potential of lncRNAs in diagnosis, prognosis and therapy in cancer, with special attention to haematological malignancies.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

