Na⁺/K⁺-ATPase β2-subunit (AMOG) expression abrogates invasion of glioblastoma-derived brain tumor-initiating cells
- PMID: 23887941
- PMCID: PMC3813412
- DOI: 10.1093/neuonc/not099
Na⁺/K⁺-ATPase β2-subunit (AMOG) expression abrogates invasion of glioblastoma-derived brain tumor-initiating cells
Abstract
Background: Mechanisms of glioma invasion remain to be fully elucidated. Glioma cells within glioblastoma multiforme (GBM) range from well-differentiated tumor cells to less-differentiated brain tumor-initiating cells (BTICs). The β2-subunit of Na(+)/K(+)-ATPase, called the adhesion molecule on glia (AMOG), is highly expressed in normal glia but is thought to be universally downregulated in GBM. To test our hypothesis that expression of AMOG is heterogeneous in GBM and confers a less invasive phenotype, we compared it between BTICs and differentiated cells from patient-matched GBM and then tested GBM invasion in vitro after AMOG overexpression.
Methods: Immunohistochemistry, immunoblotting, and real-time PCR were used to characterize AMOG protein and mRNA expression in tumor samples, BTICs, and differentiated cells. Matrigel invasion assay, scratch assay, and direct cell counting were used for testing in vitro invasion, migration, and proliferation, respectively.
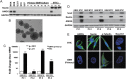
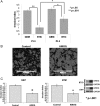
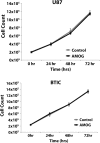
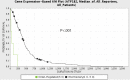
Results: While AMOG expression is heterogeneous in astrocytomas of grades II-IV, it is lost in most GBM. BTICs express higher levels of AMOG mRNA and protein compared with patient-matched differentiated tumor cells. Overexpression of AMOG decreased GBM cell and BTIC invasion without affecting migration or proliferation. Knockdown of AMOG expression in normal human astrocytes increased invasion.
Conclusions: AMOG expression inhibits GBM invasion. Its downregulation increases invasion in glial cells and may also represent an important step in BTIC differentiation. These data provide compelling evidence implicating the role of AMOG in glioma invasion and provide impetus for further investigation.
Keywords: AMOG; Na+/K+-ATPase β2-subunit; brain tumor-initiating cells; glioblastoma; invasion.
Figures



Similar articles
-
The Na, K-ATPase β-Subunit Isoforms Expression in Glioblastoma Multiforme: Moonlighting Roles.Int J Mol Sci. 2017 Nov 8;18(11):2369. doi: 10.3390/ijms18112369. Int J Mol Sci. 2017. PMID: 29117147 Free PMC article.
-
AMOG/beta2 and glioma invasion: does loss of AMOG make tumour cells run amok?Neuropathol Appl Neurobiol. 2003 Aug;29(4):370-7. doi: 10.1046/j.1365-2990.2003.00473.x. Neuropathol Appl Neurobiol. 2003. PMID: 12887597
-
Glioma malignancy is linked to interdependent and inverse AMOG and L1 adhesion molecule expression.BMC Cancer. 2019 Sep 12;19(1):911. doi: 10.1186/s12885-019-6091-5. BMC Cancer. 2019. PMID: 31510944 Free PMC article.
-
Tumor Cell Invasion in Glioblastoma.Int J Mol Sci. 2020 Mar 12;21(6):1932. doi: 10.3390/ijms21061932. Int J Mol Sci. 2020. PMID: 32178267 Free PMC article. Review.
-
Models for evaluating glioblastoma invasion along white matter tracts.Trends Biotechnol. 2024 Mar;42(3):293-309. doi: 10.1016/j.tibtech.2023.09.005. Epub 2023 Oct 6. Trends Biotechnol. 2024. PMID: 37806896 Review.
Cited by
-
Clinical significance of P-class pumps in cancer.Oncol Lett. 2021 Sep;22(3):658. doi: 10.3892/ol.2021.12919. Epub 2021 Jul 12. Oncol Lett. 2021. PMID: 34386080 Free PMC article. Review.
-
Disseminated Medulloblastoma in a Child with Germline BRCA2 6174delT Mutation and without Fanconi Anemia.Front Oncol. 2015 Aug 27;5:191. doi: 10.3389/fonc.2015.00191. eCollection 2015. Front Oncol. 2015. PMID: 26380221 Free PMC article.
-
Glycogenolysis in Acquired Glioma Resistance to Temozolomide: A Role for the [Ca2+]i-dependent Activation of Na,K-ATPase/ERK1/2 Signaling.Front Pharmacol. 2018 Aug 7;9:873. doi: 10.3389/fphar.2018.00873. eCollection 2018. Front Pharmacol. 2018. PMID: 30131700 Free PMC article.
-
The Big Picture of Glioblastoma Malignancy: A Meta-Analysis of Glioblastoma Proteomics to Identify Altered Biological Pathways.ACS Omega. 2021 Sep 14;6(38):24535-24544. doi: 10.1021/acsomega.1c02991. eCollection 2021 Sep 28. ACS Omega. 2021. PMID: 34604635 Free PMC article.
-
Na+/K+-ATPase: ion pump, signal transducer, or cytoprotective protein, and novel biological functions.Neural Regen Res. 2024 Dec 1;19(12):2684-2697. doi: 10.4103/NRR.NRR-D-23-01175. Epub 2024 Jan 31. Neural Regen Res. 2024. PMID: 38595287 Free PMC article.
References
-
- Alves TR, Lima FR, Kahn SA, et al. Glioblastoma cells: a heterogeneous and fatal tumor interacting with the parenchyma. Life Sci. 2011;89(15–16):532–539. - PubMed
-
- Parsa AT, Wachhorst S, Lamborn KR, et al. Prognostic significance of intracranial dissemination of glioblastoma multiforme in adults. J Neurosurg. 2005;102(4):622–628. - PubMed
-
- Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. - PubMed
-
- Messam CA, Hou J, Major EO. Coexpression of nestin in neural and glial cells in the developing human CNS defined by a human-specific anti-nestin antibody. Exp Neurol. 2000;161(2):585–596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

