Evaluation of genetic risk score models in the presence of interaction and linkage disequilibrium
- PMID: 23888168
- PMCID: PMC3719135
- DOI: 10.3389/fgene.2013.00138
Evaluation of genetic risk score models in the presence of interaction and linkage disequilibrium
Abstract
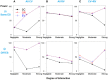
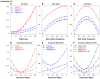
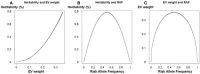
In the area of genetic epidemiology, genetic risk predictive modeling is becoming an important area of translational success. As an increasing number of genetic variants are successfully discovered, the use of multiple genetic variants in constructing a genetic risk score (GRS) for modeling has been widely applied using a variety of approaches. Previously, we compared the performance of a simple, additive GRS with weighted GRS approaches, but our initial simulation experiment assumed very simple models without many of the complications found in real genetic studies. In particular, interactions between variants and linkage disequilibrium (LD) (indirect mapping) remain important and challenging problems for GRS modeling. In the present study, we applied two simulation strategies to mimic various types of epistasis to evaluate their impact on the performance of the GRS models. We simulated a range of models demonstrating statistical interaction and linkage disequilibrium. Three genetic risk models were compared in terms of power, type I error, C-statistic and AIC, including a simple count GRS (SC-GRS), an odds ratio weighted GRS (OR-GRS) and an explained variance weighted GRS (EV-GRS). Simulation factors of interest included allele frequencies, effect sizes, strengths of interaction, degrees of LD and heritability. We extensively examined the extent to how these interactions could influence the performance of genetic risk models. Our results show that the weighted methods outperform simple count method in general even if interaction or LD is present, with well controlled type I error.
Keywords: explained variance; genetic risk score (GRS); interaction; linkage disequilibrium (LD); predictive modeling.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

