The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior
- PMID: 23889933
- PMCID: PMC3859024
- DOI: 10.1016/j.neuron.2013.05.015
The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior
Abstract
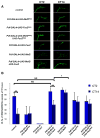
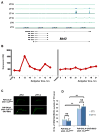
The transcription factor Mef2 regulates activity-dependent neuronal plasticity and morphology in mammals, and clock neurons are reported to experience activity-dependent circadian remodeling in Drosophila. We show here that Mef2 is required for this daily fasciculation-defasciculation cycle. Moreover, the master circadian transcription complex CLK/CYC directly regulates Mef2 transcription. ChIP-Chip analysis identified numerous Mef2 target genes implicated in neuronal plasticity, including the cell-adhesion gene Fas2. Genetic epistasis experiments support this transcriptional regulatory hierarchy, CLK/CYC- > Mef2- > Fas2, indicate that it influences the circadian fasciculation cycle within pacemaker neurons, and suggest that this cycle also contributes to circadian behavior. Mef2 therefore transmits clock information to machinery involved in neuronal remodeling, which contributes to locomotor activity rhythms.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




References
-
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008;105:9391–9396. - PMC - PubMed
-
- Behrens UD, Wagner HJ. Adaptation-dependent changes of bipolar cell terminals in fish retina: effects on overall morphology and spinule formation in Ma and Mb cells. Vision Res. 1996;36:3901–3911. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

