Subunit folds and maturation pathway of a dsRNA virus capsid
- PMID: 23891288
- PMCID: PMC3742642
- DOI: 10.1016/j.str.2013.06.007
Subunit folds and maturation pathway of a dsRNA virus capsid
Abstract
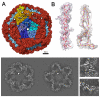
The cystovirus ϕ6 shares several distinct features with other double-stranded RNA (dsRNA) viruses, including the human pathogen, rotavirus: segmented genomes, nonequivalent packing of 120 subunits in its icosahedral capsid, and capsids as compartments for transcription and replication. ϕ6 assembles as a dodecahedral procapsid that undergoes major conformational changes as it matures into the spherical capsid. We determined the crystal structure of the capsid protein, P1, revealing a flattened trapezoid subunit with an α-helical fold. We also solved the procapsid with cryo-electron microscopy to comparable resolution. Fitting the crystal structure into the procapsid disclosed substantial conformational differences between the two P1 conformers. Maturation via two intermediate states involves remodeling on a similar scale, besides huge rigid-body rotations. The capsid structure and its stepwise maturation that is coupled to sequential packaging of three RNA segments sets the cystoviruses apart from other dsRNA viruses as a dynamic molecular machine.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures




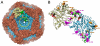
References
-
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. - PubMed
-
- Benevides JM, Juuti JT, Tuma R, Bamford DH, Thomas GJ., Jr. Characterization of subunit-specific interactions in a double-stranded RNA virus: Raman difference spectroscopy of the phi6 procapsid. Biochemistry. 2002;41:11946–11953. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

