Equine arteritis virus
- PMID: 23891306
- PMCID: PMC7126873
- DOI: 10.1016/j.vetmic.2013.06.015
Equine arteritis virus
Abstract
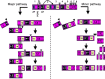
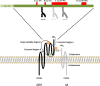
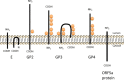
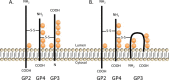
Equine arteritis virus (EAV) is the causative agent of equine viral arteritis (EVA), a respiratory and reproductive disease of equids. There has been significant recent progress in understanding the molecular biology of EAV and the pathogenesis of its infection in horses. In particular, the use of contemporary genomic techniques, along with the development and reverse genetic manipulation of infectious cDNA clones of several strains of EAV, has generated significant novel information regarding the basic molecular biology of the virus. Therefore, the objective of this review is to summarize current understanding of EAV virion architecture, replication, evolution, molecular epidemiology and genetic variation, pathogenesis including the influence of host genetics on disease susceptibility, host immune response, and potential vaccination and treatment strategies.
Keywords: EAV; EVA; Equine arteritis virus.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures






References
-
- Abes S., Moulton H.M., Clair P., Prevot P., Youngblood D.S., Wu R.P., Iversen P.L., Lebleu B. Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J. Control. Release. 2006;116:304–313. - PubMed
-
- Anonymous . Equine Viral Arteritis (EVA) and the U.S. Horse Industry. USDA-APHIS, VS, CEAH, National Animal Health Monitoring System; Fort Collins, CO: 2000. NAHMS.
-
- Anonymous . OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Mammals, Birds and Bees) 2012. 2.5.10. Equine viral arteritis.
-
- Anonymous . Press Release, Inc.; 2012. Equine Arteritis Virus cELISA Correlates Well with SN.
-
- Archambault D., St-Laurent G. Induction of apoptosis by equine arteritis virus infection. Virus Genes. 2000;20:143–147. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

