Decreased parvalbumin mRNA expression in dorsolateral prefrontal cortex in Parkinson's disease
- PMID: 23891794
- PMCID: PMC3816277
- DOI: 10.1016/j.brainres.2013.07.025
Decreased parvalbumin mRNA expression in dorsolateral prefrontal cortex in Parkinson's disease
Abstract
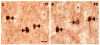
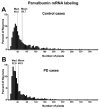
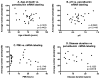
It has recently been shown that expression of the rate-limiting GABA-synthesizing enzyme glutamic acid decarboxylase (GAD) is decreased in Brodmann area 9 (BA9) of the dorsolateral prefrontal cortex (DLPFC) in Parkinson's disease (PD) compared to control brains (Lanoue, A.C., Dumitriu, A., Myers, R.H., Soghomonian, JJ., 2010. Exp. Neurol. 206 (1), 207-217). A subpopulation of cortical GABAergic interneurons expresses the calcium-binding protein parvalbumin and plays a critical role in the control of pyramidal neuron excitability and the generation of cortical gamma frequency oscillations. In view of its key role in the physiology of the cerebral cortex, we sought to determine whether the expression of parvalbumin and the number of parvalbumin-expressing neurons are altered in BA9 of PD brains. First, isotopic in situ hybridization histochemistry was used to examine mRNA expression of parvalbumin on post-mortem brain sections. Second, immunohistochemistry and design-based stereology were used to determine the density of parvalbumin-positive interneurons in BA9. Quantification of mRNA labeling at the single cell level showed a significant decrease in parvalbumin expression in PD cases. In contrast, neuronal density of parvalbumin-positive neurons was not significantly different between PD and controls. Results confirm that the GABAergic system is altered in the DLPFC in PD and identify the contribution of parvalbumin-expressing neurons in these alterations. We speculate that these effects could contribute to altered cortical excitability and oscillatory activity previously documented in PD.
Keywords: GABA; Gene expression; Parkinson′s disease; Parvalbumin; Prefrontal cortex; mRNA.
© 2013 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors have no conflict of interest to report.
Figures



References
-
- Arthur CR, Morton SL, Dunham LD, Keeney PM, Bennett JP., Jr Parkinson’s disease brain mitochondria have impaired respirasome assembly, age-related increases in distribution of oxidative damage to mtDNA and no differences in heteroplasmic mtDNA mutation abundance. Mol Neurodegener. 2009;4:37. - PMC - PubMed
-
- Bares M, Kanovsky P, Klajblova H, Rektor I. Intracortical inhibition and facilitation are impaired in patients with early Parkinson’s disease: a paired TMS study. Eur J Neurol. 2003;10:385–9. - PubMed
-
- Bartos M, Vida I, Jonas P. Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat Rev Neurosci. 2007;8:45–56. - PubMed
-
- Beasley CL, Zhang ZJ, Patten I, Reynolds GP. Selective deficits in prefrontal cortical GABAergic neurons in schizophrenia defined by the presence of calcium-binding proteins. Biol Psychiatry. 2002;52:708–15. - PubMed
-
- Benes FM, Berretta S. GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology. 2001;25:1–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

