Epigenetic signature and enhancer activity of the human APOE gene
- PMID: 23892237
- PMCID: PMC3836480
- DOI: 10.1093/hmg/ddt354
Epigenetic signature and enhancer activity of the human APOE gene
Abstract
The human apolipoprotein E (APOE) gene plays an important role in lipid metabolism. It has three common genetic variants, alleles ε2/ε3/ε4, which translate into three protein isoforms of apoE2, E3 and E4. These isoforms can differentially influence total serum cholesterol levels; therefore, APOE has been linked with cardiovascular disease. Additionally, its ε4 allele is strongly associated with the risk of Alzheimer's disease (AD), whereas the ε2 allele appears to have a modest protective effect for AD. Despite decades of research having illuminated multiple functional differences among the three apoE isoforms, the precise mechanisms through which different APOE alleles modify diseases risk remain incompletely understood. In this study, we examined the genomic structure of APOE in search for properties that may contribute novel biological consequences to the risk of disease. We identify one such element in the ε2/ε3/ε4 allele-carrying 3'-exon of APOE. We show that this exon is imbedded in a well-defined CpG island (CGI) that is highly methylated in the human postmortem brain. We demonstrate that this APOE CGI exhibits transcriptional enhancer/silencer activity. We provide evidence that this APOE CGI differentially modulates expression of genes at the APOE locus in a cell type-, DNA methylation- and ε2/ε3/ε4 allele-specific manner. These findings implicate a novel functional role for a 3'-exon CGI and support a modified mechanism of action for APOE in disease risk, involving not only the protein isoforms but also an epigenetically regulated transcriptional program at the APOE locus driven by the APOE CGI.
Figures

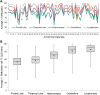



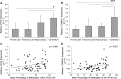
References
-
- Mahley R.W. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi:10.1126/science.3283935. - DOI - PubMed
-
- Mahley R., Huang Y. Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr. Opin. Lipidol. 1999;10:207–217. doi:10.1097/00041433-199906000-00003. - DOI - PubMed
-
- Huang Y., Weisgraber K.H., Mucke L., Mahley R.W. Apolipoprotein E: diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer's disease. J. Mol. Neurosci. 2004;23:189–204. doi:10.1385/JMN:23:3:189. - DOI - PubMed
-
- Chen Y., Lomnitski L., Michaelson D.M., Shohami E. Motor and cognitive deficits in apolipoprotein E-deficient mice after closed head injury. Neuroscience. 1997;80:1255–1262. doi:10.1016/S0306-4522(97)00007-9. - DOI - PubMed
-
- Rall S.C., Jr, Weisgraber K.H., Mahley R.W. Human apolipoprotein E. The complete amino acid sequence. J. Biol. Chem. 1982;257:4171–4178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

