Transactivation specificity is conserved among p53 family proteins and depends on a response element sequence code
- PMID: 23892287
- PMCID: PMC3794606
- DOI: 10.1093/nar/gkt657
Transactivation specificity is conserved among p53 family proteins and depends on a response element sequence code
Abstract
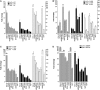
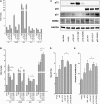
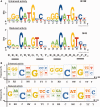
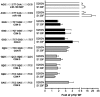
Structural and biochemical studies have demonstrated that p73, p63 and p53 recognize DNA with identical amino acids and similar binding affinity. Here, measuring transactivation activity for a large number of response elements (REs) in yeast and human cell lines, we show that p53 family proteins also have overlapping transactivation profiles. We identified mutations at conserved amino acids of loops L1 and L3 in the DNA-binding domain that tune the transactivation potential nearly equally in p73, p63 and p53. For example, the mutant S139F in p73 has higher transactivation potential towards selected REs, enhanced DNA-binding cooperativity in vitro and a flexible loop L1 as seen in the crystal structure of the protein-DNA complex. By studying, how variations in the RE sequence affect transactivation specificity, we discovered a RE-transactivation code that predicts enhanced transactivation; this correlation is stronger for promoters of genes associated with apoptosis.
Figures



References
-
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. - PubMed
-
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. - PubMed
-
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. - PubMed
-
- Collavin L, Lunardi A, Del Sal G. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ. 2010;17:901–911. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

