Carboxamide SIRT1 inhibitors block DBC1 binding via an acetylation-independent mechanism
- PMID: 23892437
- PMCID: PMC3755073
- DOI: 10.4161/cc.25268
Carboxamide SIRT1 inhibitors block DBC1 binding via an acetylation-independent mechanism
Abstract
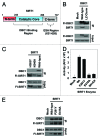
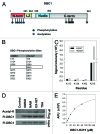
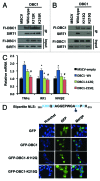
SIRT1 is an NAD (+) -dependent deacetylase that counteracts multiple disease states associated with aging and may underlie some of the health benefits of calorie restriction. Understanding how SIRT1 is regulated in vivo could therefore lead to new strategies to treat age-related diseases. SIRT1 forms a stable complex with DBC1, an endogenous inhibitor. Little is known regarding the biochemical nature of SIRT1-DBC1 complex formation, how it is regulated and whether or not it is possible to block this interaction pharmacologically. In this study, we show that critical residues within the catalytic core of SIRT1 mediate binding to DBC1 via its N-terminal region, and that several carboxamide SIRT1 inhibitors, including EX-527, can completely block this interaction. We identify two acetylation sites on DBC1 that regulate its ability to bind SIRT1 and suppress its activity. Furthermore, we show that DBC1 itself is a substrate for SIRT1. Surprisingly, the effect of EX-527 on SIRT1-DBC1 binding is independent of DBC1 acetylation. Together, these data show that protein acetylation serves as an endogenous regulatory mechanism for SIRT1-DBC1 binding and illuminate a new path to developing small-molecule modulators of SIRT1.
Keywords: DBC1; DBC1 acetylation; DBC1 localization; SIRT1 inhibitors; SIRT1-DBC1 complex regulation.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
