Comparative pharmacology of bombesin receptor subtype-3, nonpeptide agonist MK-5046, a universal peptide agonist, and peptide antagonist Bantag-1 for human bombesin receptors
- PMID: 23892571
- PMCID: PMC3781414
- DOI: 10.1124/jpet.113.206896
Comparative pharmacology of bombesin receptor subtype-3, nonpeptide agonist MK-5046, a universal peptide agonist, and peptide antagonist Bantag-1 for human bombesin receptors
Abstract
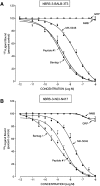
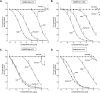
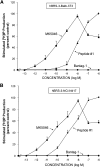
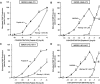
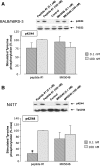
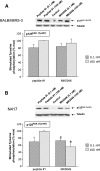
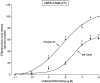
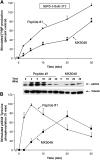
Bombesin-receptor-subtype-3 (BRS-3) is an orphan G-protein-coupled receptor of the bombesin (Bn) family whose natural ligand is unknown and which does not bind any natural Bn-peptide with high affinity. It is present in the central nervous system, peripheral tissues, and tumors; however, its role in normal physiology/pathophysiology is largely unknown because of the lack of selective ligands. Recently, MK-5046 [(2S)-1,1,1-trifluoro-2-[4-(1H-pyrazol-1-yl)phenyl]-3-(4-{[1-(trifluoromethyl)cyclopropyl]methyl}-1H-imidazol-2-yl)propan-2-ol] and Bantag-1 [Boc-Phe-His-4-amino-5-cyclohexyl-2,4,5-trideoxypentonyl-Leu-(3-dimethylamino) benzylamide N-methylammonium trifluoroacetate], a nonpeptide agonist and a peptide antagonist, respectively, for BRS-3 have been described, but there have been limited studies on their pharmacology. We studied MK-5046 and Bantag-1 interactions with human Bn-receptors-human bombesin receptor subtype-3 (hBRS-3), gastrin-releasing peptide receptor (GRP-R), and neuromedin B receptor (NMB-R)-and compared them with the nonselective, peptide-agonist [d-Tyr6,βAla11,Phe13,Nle14]Bn-(6-14) (peptide #1). Receptor activation was detected by activation of phospholipase C (PLC), mitogen-activated protein kinase (MAPK), focal adhesion kinase (FAK), paxillin, and Akt. In hBRS-3 cells, the relative affinities were Bantag-1 (1.3 nM) > peptide #1 (2 nM) > MK-5046 (37-160 nM) > GRP, NMB (>10 μM), and the binding-dose-inhibition curves were broad (>4 logs), with Hill coefficients differing significantly from unity. Curve-fitting demonstrated high-affinity (MK-5046, Ki = 0.08 nM) and low-affinity (MK-5046, Ki = 11-29 nM) binding sites. For PLC activation in hBRS-3 cells, the relative potencies were MK-5046 (0.02 nM) > peptide #1 (6 nM) > GRP, NMB, Bantag-1 (>10 μM), and MK-5046 had a biphasic dose response, whereas peptide #1 was monophasic. Bantag-1 was a specific hBRS-3-antagonist. In hBRS-3 cells, MK-5046 was a full agonist for activation of MAPK, FAK, Akt, and paxillin; however, it was a partial agonist for phospholipase A2 (PLA2) activation. The kinetics of activation/duration of action for PLC/MAPK activation of MK-5046 and peptide #1 differed, with peptide #1 causing more rapid stimulation; however, MK-5046 had more prolonged activity. Our study finds that MK-5046 and Bantag-1 have high affinity/selectivity for hBRS-3. The nonpeptide MK-5046 and peptide #1 agonists differ markedly in their receptor coupling, ability to activate different signaling cascades, and kinetics/duration of action. These results show that their hBRS-3 receptor activation is not always concordant and could lead to markedly different cellular responses.
Figures




References
-
- Aramori I, Zenkoh J, Morikawa N, Asano M, Hatori C, Sawai H, Kayakiri H, Satoh S, Inoue T, Abe Y, et al. (1997) Nonpeptide mimic of bradykinin with long-acting properties at the bradykinin B2 receptor. Mol Pharmacol 52:16–20 - PubMed
-
- Benya RV, Fathi Z, Kusui T, Pradhan T, Battey JF, Jensen RT. (1994) Gastrin-releasing peptide receptor-induced internalization, down-regulation, desensitization, and growth: possible role for cyclic AMP. Mol Pharmacol 46:235–245 - PubMed
-
- Benya RV, Kusui T, Pradhan TK, Battey JF, Jensen RT. (1995) Expression and characterization of cloned human bombesin receptors. Mol Pharmacol 47:10–20 - PubMed
-
- Benya RV, Wada E, Battey JF, Fathi Z, Wang LH, Mantey SA, Coy DH, Jensen RT. (1992) Neuromedin B receptors retain functional expression when transfected into BALB 3T3 fibroblasts: analysis of binding, kinetics, stoichiometry, modulation by guanine nucleotide-binding proteins, and signal transduction and comparison with natively expressed receptors. Mol Pharmacol 42:1058–1068 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

