Distinct amino acids of histone H3 control secondary metabolism in Aspergillus nidulans
- PMID: 23892751
- PMCID: PMC3811361
- DOI: 10.1128/AEM.01578-13
Distinct amino acids of histone H3 control secondary metabolism in Aspergillus nidulans
Abstract
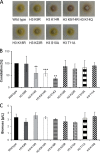
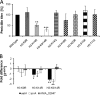
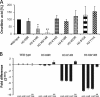
Chromatin remodelling events play an important role in the secondary metabolism of filamentous fungi. Previously, we showed that a bacterium, Streptomyces rapamycinicus, is able to reprogram the histone-modifying Spt-Ada-Gcn5-acetyltransferase/ADA (SAGA/ADA) complex of the model fungus Aspergillus nidulans. Consequently, the histone H3 amino acids lysine 9 and lysine 14 at distinct secondary metabolism genes were specifically acetylated during the bacterial fungal interaction, which, furthermore, was associated with the activation of the otherwise silent orsellinic acid gene cluster. To investigate the importance of the histone modifications for distinct gene expression profiles in fungal secondary metabolism, we exchanged several amino acids of histone H3 of A. nidulans. These amino acids included lysine residues 9, 14, 18, and 23 as well as serine 10 and threonine 11. Lysine residues were replaced by arginine or glutamine residues, and serine/threonine residues were replaced by alanine. All generated mutant strains were viable, allowing direct analysis of the consequences of missing posttranslational histone modifications. In the mutant strains, major changes in the expression patterns at both the transcriptional and metabolite levels of the penicillin, sterigmatocystin, and orsellinic acid biosynthesis gene clusters were detected. These effects were due mainly to the substitution of the acetylatable lysine 14 of histone H3 and were enhanced in a lysine 14/lysine 9 double mutant of histone H3. Taken together, our findings show a causal linkage between the acetylation of lysine residue 14 of histone H3 and the transcription and product formation of secondary metabolite gene clusters.
Figures


Similar articles
-
Chromatin mapping identifies BasR, a key regulator of bacteria-triggered production of fungal secondary metabolites.Elife. 2018 Oct 12;7:e40969. doi: 10.7554/eLife.40969. Elife. 2018. PMID: 30311911 Free PMC article.
-
Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation.Proc Natl Acad Sci U S A. 2011 Aug 23;108(34):14282-7. doi: 10.1073/pnas.1103523108. Epub 2011 Aug 8. Proc Natl Acad Sci U S A. 2011. PMID: 21825172 Free PMC article.
-
KdmB, a Jumonji Histone H3 Demethylase, Regulates Genome-Wide H3K4 Trimethylation and Is Required for Normal Induction of Secondary Metabolism in Aspergillus nidulans.PLoS Genet. 2016 Aug 22;12(8):e1006222. doi: 10.1371/journal.pgen.1006222. eCollection 2016 Aug. PLoS Genet. 2016. PMID: 27548260 Free PMC article.
-
Coordinate control of secondary metabolite production and asexual sporulation in Aspergillus nidulans.Curr Opin Microbiol. 1998 Dec;1(6):674-7. doi: 10.1016/s1369-5274(98)80114-8. Curr Opin Microbiol. 1998. PMID: 10066549 Review.
-
Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans.J Ind Microbiol Biotechnol. 2014 Feb;41(2):433-42. doi: 10.1007/s10295-013-1386-z. Epub 2013 Dec 17. J Ind Microbiol Biotechnol. 2014. PMID: 24342965 Free PMC article. Review.
Cited by
-
Chromatin Manipulation and Editing: Challenges, New Technologies and Their Use in Plants.Int J Mol Sci. 2021 Jan 6;22(2):512. doi: 10.3390/ijms22020512. Int J Mol Sci. 2021. PMID: 33419220 Free PMC article. Review.
-
Strategies for mining fungal natural products.J Ind Microbiol Biotechnol. 2014 Feb;41(2):301-13. doi: 10.1007/s10295-013-1366-3. Epub 2013 Oct 22. J Ind Microbiol Biotechnol. 2014. PMID: 24146366 Review.
-
The Aspergillus flavus Histone Acetyltransferase AflGcnE Regulates Morphogenesis, Aflatoxin Biosynthesis, and Pathogenicity.Front Microbiol. 2016 Aug 30;7:1324. doi: 10.3389/fmicb.2016.01324. eCollection 2016. Front Microbiol. 2016. PMID: 27625637 Free PMC article.
-
Pestalotiopsis Diversity: Species, Dispositions, Secondary Metabolites, and Bioactivities.Molecules. 2022 Nov 21;27(22):8088. doi: 10.3390/molecules27228088. Molecules. 2022. PMID: 36432188 Free PMC article. Review.
-
Chromatin mapping identifies BasR, a key regulator of bacteria-triggered production of fungal secondary metabolites.Elife. 2018 Oct 12;7:e40969. doi: 10.7554/eLife.40969. Elife. 2018. PMID: 30311911 Free PMC article.
References
-
- Brakhage AA. 2013. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11:21–32 - PubMed
-
- Bergmann S, Funk AN, Scherlach K, Schroeckh V, Shelest E, Horn U, Hertweck C, Brakhage AA. 2010. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl. Environ. Microbiol. 76:8143–8149 - PMC - PubMed
-
- Bergmann S, Schuemann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. 2007. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 3:213–217 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

