Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA
- PMID: 23892783
- PMCID: PMC3808885
- DOI: 10.1038/nature12440
Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA
Abstract
In Gram-positive bacteria, T-box riboswitches regulate the expression of aminoacyl-tRNA synthetases and other proteins in response to fluctuating transfer RNA aminoacylation levels under various nutritional states. T-boxes reside in the 5'-untranslated regions of the messenger RNAs they regulate, and consist of two conserved domains. Stem I contains the specifier trinucleotide that base pairs with the anticodon of cognate tRNA. 3' to stem I is the antiterminator domain, which base pairs with the tRNA acceptor end and evaluates its aminoacylation state. Despite high phylogenetic conservation and widespread occurrence in pathogens, the structural basis of tRNA recognition by this riboswitch remains ill defined. Here we demonstrate that the ~100-nucleotide T-box stem I is necessary and sufficient for specific, high-affinity (dissociation constant (Kd) ~150 nM) tRNA binding, and report the structure of Oceanobacillus iheyensis glyQ stem I in complex with its cognate tRNA at 3.2 Å resolution. Stem I recognizes the overall architecture of tRNA in addition to its anticodon, something accomplished by large ribonucleoproteins such as the ribosome, or proteins such as aminoacyl-tRNA synthetases, but is unprecedented for a compact mRNA domain. The C-shaped stem I cradles the L-shaped tRNA, forming an extended (1,604 Å(2)) intermolecular interface. In addition to the specifier-anticodon interaction, two interdigitated T-loops near the apex of stem I stack on the tRNA elbow in a manner analogous to those of the J11/12-J12/11 motif of RNase P and the L1 stalk of the ribosomal E-site. Because these ribonucleoproteins and T-boxes are unrelated, this strategy to recognize a universal tRNA feature probably evolved convergently. Mutually induced fit of stem I and the tRNA exploiting the intrinsic flexibility of tRNA and its conserved post-transcriptional modifications results in high shape complementarity, which in addition to providing specificity and affinity, globally organizes the T-box to orchestrate tRNA-dependent transcription regulation.
Figures
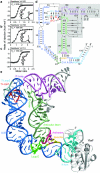
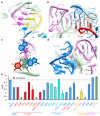
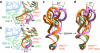
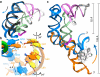
Comment in
-
Structural biology: RNA exerts self-control.Nature. 2013 Aug 15;500(7462):279-80. doi: 10.1038/nature12460. Epub 2013 Jul 28. Nature. 2013. PMID: 23892780 Free PMC article.
Similar articles
-
Capture and Release of tRNA by the T-Loop Receptor in the Function of the T-Box Riboswitch.Biochemistry. 2017 Jul 18;56(28):3549-3558. doi: 10.1021/acs.biochem.7b00284. Epub 2017 Jul 3. Biochemistry. 2017. PMID: 28621923 Free PMC article.
-
An evolving tale of two interacting RNAs-themes and variations of the T-box riboswitch mechanism.IUBMB Life. 2019 Aug;71(8):1167-1180. doi: 10.1002/iub.2098. Epub 2019 Jun 17. IUBMB Life. 2019. PMID: 31206978 Free PMC article. Review.
-
Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout.Mol Cell. 2014 Jul 3;55(1):148-55. doi: 10.1016/j.molcel.2014.05.017. Epub 2014 Jun 19. Mol Cell. 2014. PMID: 24954903 Free PMC article.
-
Solution structure of the K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-box leader RNA.J Mol Biol. 2011 Apr 22;408(1):99-117. doi: 10.1016/j.jmb.2011.02.014. Epub 2011 Feb 17. J Mol Biol. 2011. PMID: 21333656 Free PMC article.
-
Structure and mechanism of the T-box riboswitches.Wiley Interdiscip Rev RNA. 2015 Jul-Aug;6(4):419-33. doi: 10.1002/wrna.1285. Epub 2015 May 8. Wiley Interdiscip Rev RNA. 2015. PMID: 25959893 Free PMC article. Review.
Cited by
-
Structural Insights into RNA Dimerization: Motifs, Interfaces and Functions.Molecules. 2020 Jun 23;25(12):2881. doi: 10.3390/molecules25122881. Molecules. 2020. PMID: 32585844 Free PMC article. Review.
-
An aminoacylation ribozyme evolved from a natural tRNA-sensing T-box riboswitch.Nat Chem Biol. 2020 Jun;16(6):702-709. doi: 10.1038/s41589-020-0500-6. Epub 2020 Mar 23. Nat Chem Biol. 2020. PMID: 32203413
-
RNA cis-regulators are important for Streptococcus pneumoniae in vivo success.PLoS Genet. 2024 Mar 5;20(3):e1011188. doi: 10.1371/journal.pgen.1011188. eCollection 2024 Mar. PLoS Genet. 2024. PMID: 38442125 Free PMC article.
-
Interplay between Host tRNAs and HIV-1: A Structural Perspective.Viruses. 2021 Sep 13;13(9):1819. doi: 10.3390/v13091819. Viruses. 2021. PMID: 34578400 Free PMC article. Review.
-
Unique anticodon loop conformation with the flipped-out wobble nucleotide in the crystal structure of unbound tRNAVal.RNA. 2021 Nov;27(11):1330-1338. doi: 10.1261/rna.078863.121. Epub 2021 Jul 27. RNA. 2021. PMID: 34315814 Free PMC article.
References
-
- Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. - PubMed
-
- Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. J. Mol. Biol. 2005;349:273–287. - PubMed
-
- Perona JJ, Hadd A. Structural diversity and protein engineering of the aminoacyl-tRNA synthetases. Biochemistry. 2012;51:8705–8729. - PubMed
Methods and Extended Data References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

