The core microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a nonspecific RNA-binding protein
- PMID: 23893406
- PMCID: PMC3772224
- DOI: 10.1074/jbc.M112.446880
The core microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a nonspecific RNA-binding protein
Abstract
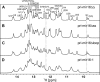
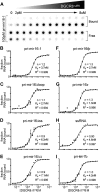
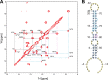
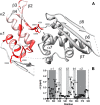
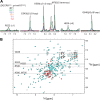
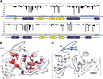
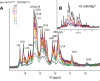
MicroRNA (miRNA) biogenesis follows a conserved succession of processing steps, beginning with the recognition and liberation of an miRNA-containing precursor miRNA hairpin from a large primary miRNA transcript (pri-miRNA) by the Microprocessor, which consists of the nuclear RNase III Drosha and the double-stranded RNA-binding domain protein DGCR8 (DiGeorge syndrome critical region protein 8). Current models suggest that specific recognition is driven by DGCR8 detection of single-stranded elements of the pri-miRNA stem-loop followed by Drosha recruitment and pri-miRNA cleavage. Because countless RNA transcripts feature single-stranded-dsRNA junctions and DGCR8 can bind hundreds of mRNAs, we explored correlations between RNA binding properties of DGCR8 and specific pri-miRNA substrate processing. We found that DGCR8 bound single-stranded, double-stranded, and random hairpin transcripts with similar affinity. Further investigation of DGCR8/pri-mir-16 interactions by NMR detected intermediate exchange regimes over a wide range of stoichiometric ratios. Diffusion analysis of DGCR8/pri-mir-16 interactions by pulsed field gradient NMR lent further support to dynamic complex formation involving free components in exchange with complexes of varying stoichiometry, although in vitro processing assays showed exclusive cleavage of pri-mir-16 variants bearing single-stranded flanking regions. Our results indicate that DGCR8 binds RNA nonspecifically. Therefore, a sequential model of DGCR8 recognition followed by Drosha recruitment is unlikely. Known RNA substrate requirements are broad and include 70-nucleotide hairpins with unpaired flanking regions. Thus, specific RNA processing is likely facilitated by preformed DGCR8-Drosha heterodimers that can discriminate between authentic substrates and other hairpins.
Keywords: DGCR8; Double-stranded RNA-binding Domain (dsRBD); Drosha; MicroRNA; Microprocessor; NMR; Protein-Nucleic Acid Interaction; RNA Structure; RNA-binding Protein.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

