Ras-mutant cancer cells display B-Raf binding to Ras that activates extracellular signal-regulated kinase and is inhibited by protein kinase A phosphorylation
- PMID: 23893412
- PMCID: PMC3779760
- DOI: 10.1074/jbc.M113.463067
Ras-mutant cancer cells display B-Raf binding to Ras that activates extracellular signal-regulated kinase and is inhibited by protein kinase A phosphorylation
Abstract
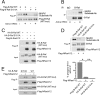
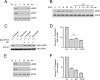
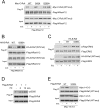
The small G protein Ras regulates proliferation through activation of the mitogen-activated protein (MAP) kinase (ERK) cascade. The first step of Ras-dependent activation of ERK signaling is Ras binding to members of the Raf family of MAP kinase kinase kinases, C-Raf and B-Raf. Recently, it has been reported that in melanoma cells harboring oncogenic Ras mutations, B-Raf does not bind to Ras and does not contribute to basal ERK activation. For other types of Ras-mutant tumors, the relative contributions of C-Raf and B-Raf are not known. We examined non-melanoma cancer cell lines containing oncogenic Ras mutations and express both C-Raf and B-Raf isoforms, including the lung cancer cell line H1299 cells. Both B-Raf and C-Raf were constitutively bound to oncogenic Ras and contributed to Ras-dependent ERK activation. Ras binding to B-Raf and C-Raf were both subject to inhibition by the cAMP-dependent protein kinase PKA. cAMP inhibited the growth of H1299 cells and Ras-dependent ERK activation via PKA. PKA inhibited the binding of Ras to both C-Raf and B-Raf through phosphorylations of C-Raf at Ser-259 and B-Raf at Ser-365, respectively. These studies demonstrate that in non-melanocytic Ras-mutant cancer cells, Ras signaling to B-Raf is a significant contributor to ERK activation and that the B-Raf pathway, like that of C-Raf, is a target for inhibition by PKA. We suggest that cAMP and hormones coupled to cAMP may prove useful in dampening the effects of oncogenic Ras in non-melanocytic cancer cells through PKA-dependent actions on B-Raf as well as C-Raf.
Keywords: B-Raf; C-Raf; ERK; MAP Kinases (MAPKs); Protein Kinase A (PKA); Raf; Ras.
Figures





Similar articles
-
Phosphorylation of Rap1 by cAMP-dependent Protein Kinase (PKA) Creates a Binding Site for KSR to Sustain ERK Activation by cAMP.J Biol Chem. 2017 Jan 27;292(4):1449-1461. doi: 10.1074/jbc.M116.768986. Epub 2016 Dec 21. J Biol Chem. 2017. PMID: 28003362 Free PMC article.
-
Protein Kinase A-independent Ras Protein Activation Cooperates with Rap1 Protein to Mediate Activation of the Extracellular Signal-regulated Kinases (ERK) by cAMP.J Biol Chem. 2016 Oct 7;291(41):21584-21595. doi: 10.1074/jbc.M116.730978. Epub 2016 Aug 16. J Biol Chem. 2016. PMID: 27531745 Free PMC article.
-
Cyclic AMP signaling reduces sirtuin 6 expression in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK (Raf/mitogen-activated extracellular signal-regulated kinase/extracellular signal-regulated kinase) pathway.J Biol Chem. 2015 Apr 10;290(15):9604-13. doi: 10.1074/jbc.M114.633198. Epub 2015 Feb 24. J Biol Chem. 2015. PMID: 25713071 Free PMC article.
-
RAF protein-serine/threonine kinases: structure and regulation.Biochem Biophys Res Commun. 2010 Aug 27;399(3):313-7. doi: 10.1016/j.bbrc.2010.07.092. Epub 2010 Jul 30. Biochem Biophys Res Commun. 2010. PMID: 20674547 Review.
-
RAF-MEK-ERK pathway in cancer evolution and treatment.Semin Cancer Biol. 2022 Oct;85:123-154. doi: 10.1016/j.semcancer.2021.05.010. Epub 2021 May 13. Semin Cancer Biol. 2022. PMID: 33992782 Review.
Cited by
-
Disruption of the pro-oncogenic c-RAF-PDE8A complex represents a differentiated approach to treating KRAS-c-RAF dependent PDAC.Sci Rep. 2024 Apr 18;14(1):8998. doi: 10.1038/s41598-024-59451-3. Sci Rep. 2024. PMID: 38637546 Free PMC article.
-
Phosphorylation of the C-Raf N Region Promotes Raf Dimerization.Mol Cell Biol. 2017 Sep 12;37(19):e00132-17. doi: 10.1128/MCB.00132-17. Print 2017 Oct 1. Mol Cell Biol. 2017. PMID: 28694330 Free PMC article.
-
The Role of Anti-EGFR Monoclonal Antibody in mCRC Maintenance Therapy.Front Mol Biosci. 2022 Mar 30;9:870395. doi: 10.3389/fmolb.2022.870395. eCollection 2022. Front Mol Biosci. 2022. PMID: 35433839 Free PMC article. Review.
-
The metabolic/pH sensor soluble adenylyl cyclase is a tumor suppressor protein.Oncotarget. 2016 Jul 19;7(29):45597-45607. doi: 10.18632/oncotarget.10056. Oncotarget. 2016. PMID: 27323809 Free PMC article.
-
Phosphorylation of Rap1 by cAMP-dependent Protein Kinase (PKA) Creates a Binding Site for KSR to Sustain ERK Activation by cAMP.J Biol Chem. 2017 Jan 27;292(4):1449-1461. doi: 10.1074/jbc.M116.768986. Epub 2016 Dec 21. J Biol Chem. 2017. PMID: 28003362 Free PMC article.
References
-
- Bos J. L. (1989) ras oncogenes in human cancer. A review. Cancer Res. 49, 4682–4689 - PubMed
-
- Rinehart J., Adjei A. A., Lorusso P. M., Waterhouse D., Hecht J. R., Natale R. B., Hamid O., Varterasian M., Asbury P., Kaldjian E. P., Gulyas S., Mitchell D. Y., Herrera R., Sebolt-Leopold J. S., Meyer M. B. (2004) Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J. Clin. Oncol. 22, 4456–4462 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

