β-Hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40 S ribosome binding to regulate initiator tRNA(Met) recruitment and accuracy of AUG selection in vivo
- PMID: 23893413
- PMCID: PMC3779751
- DOI: 10.1074/jbc.M113.498642
β-Hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40 S ribosome binding to regulate initiator tRNA(Met) recruitment and accuracy of AUG selection in vivo
Abstract
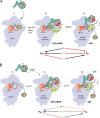
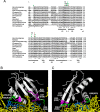
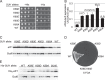
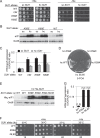
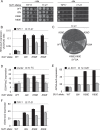
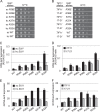
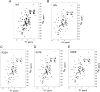
Recognition of the translation initiation codon is thought to require dissociation of eIF1 from the 40 S ribosomal subunit, enabling irreversible GTP hydrolysis (Pi release) by the eIF2·GTP·Met-tRNAi ternary complex (TC), rearrangement of the 40 S subunit to a closed conformation incompatible with scanning, and stable binding of Met-tRNAi to the P site. The crystal structure of a Tetrahymena 40 S·eIF1 complex revealed several basic amino acids in eIF1 contacting 18 S rRNA, and we tested the prediction that their counterparts in yeast eIF1 are required to prevent premature eIF1 dissociation from scanning ribosomes at non-AUG triplets. Supporting this idea, substituting Lys-60 in helix α1, or either Lys-37 or Arg-33 in β-hairpin loop-1, impairs binding of yeast eIF1 to 40 S·eIF1A complexes in vitro, and it confers increased initiation at UUG codons (Sui(-) phenotype) or lethality, in a manner suppressed by overexpressing the mutant proteins or by an eIF1A mutation (17-21) known to impede eIF1 dissociation in vitro. The eIF1 Sui(-) mutations also derepress translation of GCN4 mRNA, indicating impaired ternary complex loading, and this Gcd(-) phenotype is likewise suppressed by eIF1 overexpression or the 17-21 mutation. These findings indicate that direct contacts of eIF1 with 18 S rRNA seen in the Tetrahymena 40 S·eIF1 complex are crucial in yeast to stabilize the open conformation of the 40 S subunit and are required for rapid TC loading and ribosomal scanning and to impede rearrangement to the closed complex at non-AUG codons. Finally, we implicate the unstructured N-terminal tail of eIF1 in blocking rearrangement to the closed conformation in the scanning preinitiation complex.
Keywords: AUG Recognition; Ribosome Function; Ribosomes; Transfer RNA (tRNA); Translation; Translation Initiation Factors; eIF1; eIF2.
Figures



References
-
- Pestova T. V., Lorsch J. R., Hellen C. U. T. (2007) in Translational Control in Biology and Medicine (Mathews M., Sonenberg N., Hershey J. W., eds) pp. 87–128, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
-
- Algire M. A., Maag D., Lorsch J. R. (2005) Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20, 251–262 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

